Mapping the diverse structural landscape of the flavivirus antibody repertoire
- PMID: 32801077
- PMCID: PMC7746604
- DOI: 10.1016/j.coviro.2020.07.006
Mapping the diverse structural landscape of the flavivirus antibody repertoire
Abstract
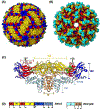
Flaviviruses are emerging arthropod-borne RNA viruses, causing a broad spectrum of life-threatening disease symptoms such as encephalitis and hemorrhagic fever. Successful vaccines exist against yellow fever virus, Japanese encephalitis virus and tick-borne encephalitis virus. However, vaccine development against other flaviviruses like dengue virus is not straightforward. This is partly because of the high sequence conservation and immunological cross-reactivity among flavivirus envelope glycoproteins leading to antibody mediated enhancement of disease. A comprehensive analyses of the structural landscape of humoral immune response against flaviviruses is crucial for antigen design. Here, we compare the available structural data of several flavivirus antibody complexes with a major focus on Zika virus and dengue virus and discuss the mapped epitopes, the stoichiometry of antibody binding and mechanisms of neutralization.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of Interest Statement
The authors declare that there is no conflict of interest.
Figures



References
-
- Lindenbach BD, Rice CM: Molecular biology of flaviviruses. Adv. Virus Res 2003, 59:23–61. - PubMed
-
- Guzman MG, Harris E: Dengue. Lancet 2015, 385:453–465. - PubMed
-
- Butler D: Fears rise over yellow fever’s next move. Nature 2016, 532:155–156. - PubMed
-
- Sejvar JJ: West Nile virus infection. Microbiol Spectr. 2016, 4:EI10–0021-2016. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

