Detection and characterization of jagged ends of double-stranded DNA in plasma
- PMID: 32801148
- PMCID: PMC7462074
- DOI: 10.1101/gr.261396.120
Detection and characterization of jagged ends of double-stranded DNA in plasma
Abstract
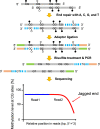
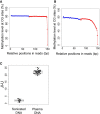
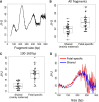
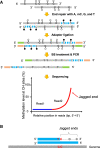
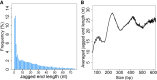
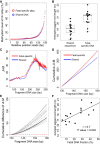
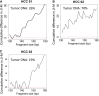
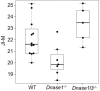
Cell-free DNA in plasma has been used for noninvasive prenatal testing and cancer liquid biopsy. The physical properties of cell-free DNA fragments in plasma, such as fragment sizes and ends, have attracted much recent interest, leading to the emerging field of cell-free DNA fragmentomics. However, one aspect of plasma DNA fragmentomics as to whether double-stranded plasma molecules might carry single-stranded ends, termed a jagged end in this study, remains underexplored. We have developed two approaches for investigating the presence of jagged ends in a plasma DNA pool. These approaches utilized DNA end repair to introduce differential methylation signals between the original sequence and the jagged ends, depending on whether unmethylated or methylated cytosines were used in the DNA end-repair procedure. The majority of plasma DNA molecules (87.8%) were found to bear jagged ends. The jaggedness varied according to plasma DNA fragment sizes and appeared to be in association with nucleosomal patterns. In the plasma of pregnant women, the jaggedness of fetal DNA molecules was higher than that of the maternal counterparts. The jaggedness of plasma DNA correlated with the fetal DNA fraction. Similarly, in the plasma of cancer patients, tumor-derived DNA molecules in patients with hepatocellular carcinoma showed an elevated jaggedness compared with nontumoral DNA. In mouse models, knocking out of the Dnase1 gene reduced jaggedness, whereas knocking out of the Dnase1l3 gene enhanced jaggedness. Hence, plasma DNA jagged ends represent an intrinsic property of plasma DNA and provide a link between nuclease activities and the fragmentation of plasma DNA.
© 2020 Jiang et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

Similar articles
-
Jagged Ends on Multinucleosomal Cell-Free DNA Serve as a Biomarker for Nuclease Activity and Systemic Lupus Erythematosus.Clin Chem. 2022 Jul 3;68(7):917-926. doi: 10.1093/clinchem/hvac050. Clin Chem. 2022. PMID: 35587043
-
Jagged Ends of Urinary Cell-Free DNA: Characterization and Feasibility Assessment in Bladder Cancer Detection.Clin Chem. 2021 Mar 31;67(4):621-630. doi: 10.1093/clinchem/hvaa325. Clin Chem. 2021. PMID: 33604652
-
Fragmentomics of urinary cell-free DNA in nuclease knockout mouse models.PLoS Genet. 2022 Jul 6;18(7):e1010262. doi: 10.1371/journal.pgen.1010262. eCollection 2022 Jul. PLoS Genet. 2022. PMID: 35793278 Free PMC article.
-
Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies.Science. 2021 Apr 9;372(6538):eaaw3616. doi: 10.1126/science.aaw3616. Science. 2021. PMID: 33833097 Review.
-
Cell-Free DNA Fragmentomics in Liquid Biopsy.Diagnostics (Basel). 2022 Apr 13;12(4):978. doi: 10.3390/diagnostics12040978. Diagnostics (Basel). 2022. PMID: 35454026 Free PMC article. Review.
Cited by
-
Mining nucleic acid "omics" to boost liquid biopsy in cancer.Cell Rep Med. 2024 Sep 17;5(9):101736. doi: 10.1016/j.xcrm.2024.101736. Cell Rep Med. 2024. PMID: 39293399 Free PMC article. Review.
-
Multi-modal cell-free DNA genomic and fragmentomic patterns enhance cancer survival and recurrence analysis.Cell Rep Med. 2024 Jan 16;5(1):101349. doi: 10.1016/j.xcrm.2023.101349. Epub 2023 Dec 20. Cell Rep Med. 2024. PMID: 38128532 Free PMC article.
-
FinaleMe: Predicting DNA methylation by the fragmentation patterns of plasma cell-free DNA.Nat Commun. 2024 Mar 30;15(1):2790. doi: 10.1038/s41467-024-47196-6. Nat Commun. 2024. PMID: 38555308 Free PMC article.
-
Genomic approaches to cancer and minimal residual disease detection using circulating tumor DNA.J Immunother Cancer. 2023 Jun;11(6):e006284. doi: 10.1136/jitc-2022-006284. J Immunother Cancer. 2023. PMID: 37349125 Free PMC article. Review.
-
A clinician's handbook for using ctDNA throughout the patient journey.Mol Cancer. 2022 Mar 21;21(1):81. doi: 10.1186/s12943-022-01551-7. Mol Cancer. 2022. PMID: 35307037 Free PMC article. Review.
References
-
- Chan KCA, Jiang P, Sun K, Cheng YKY, Tong YK, Cheng SH, Wong AIC, Hudecova I, Leung TY, Chiu RWK, et al. 2016. Second generation noninvasive fetal genome analysis reveals de novo mutations, single-base parental inheritance, and preferred DNA ends. Proc Natl Acad Sci 113: E8159–E8168. 10.1073/pnas.1615800113 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
