Triggering a switch from basal- to luminal-like breast cancer subtype by the small-molecule diptoindonesin G via induction of GABARAPL1
- PMID: 32801338
- PMCID: PMC7429843
- DOI: 10.1038/s41419-020-02878-z
Triggering a switch from basal- to luminal-like breast cancer subtype by the small-molecule diptoindonesin G via induction of GABARAPL1
Abstract
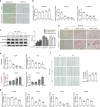
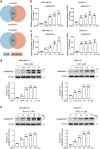
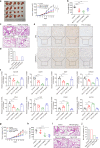
Breast cancer is a heterogeneous disease that includes different molecular subtypes. The basal-like subtype has a poor prognosis and a high recurrence rate, whereas the luminal-like subtype confers a more favorable patient prognosis partially due to anti-hormone therapy responsiveness. Here, we demonstrate that diptoindonesin G (Dip G), a natural product, exhibits robust differentiation-inducing activity in basal-like breast cancer cell lines and animal models. Specifically, Dip G treatment caused a partial transcriptome shift from basal to luminal gene expression signatures and prompted sensitization of basal-like breast tumors to tamoxifen therapy. Dip G upregulated the expression of both GABARAPL1 (GABAA receptor-associated protein-like 1) and ERβ. We revealed a previously unappreciated role of GABARAPL1 as a regulator in the specification of breast cancer subtypes that is dependent on ERβ levels. Our findings shed light on new therapeutic opportunities for basal-like breast cancer via a phenotype switch and indicate that Dip G may serve as a leading compound for the therapy of basal-like breast cancer.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
-
- Early Breast Cancer Trialists’ Collaborative, G. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. - PubMed
-
- Voduc KD, et al. Breast cancer subtypes and the risk of local and regional relapse. J. Clin. Oncol. 2010;28:1684–1691. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

