Genetic diversity and population structure of endangered plant species Anagallis foemina Mill. [ Lysimachia foemina (Mill.) U. Manns & Anderb.]
- PMID: 32801495
- PMCID: PMC7415047
- DOI: 10.1007/s12298-020-00839-6
Genetic diversity and population structure of endangered plant species Anagallis foemina Mill. [ Lysimachia foemina (Mill.) U. Manns & Anderb.]
Abstract
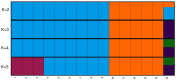
Anagallis foemina L. [syn. Lysimachia foemina (Mill.) U. Manns & Anderb.] is an annual, segetal weed from the family Primulaceae, recognized as a very rare and endangered species in many European countries. The rare occurrence of this species is associated with the specificity of the habitats in which it occurs. Knowledge of genetic diversity within and between rare species populations is a crucial step for investigating the causes of extinction as well as developing effective conservation strategies. The current study undertakes the assessment of the genetic variation and population structure of Anagallis foemina L. specimens collected in south-eastern Poland, Volhynian Polesie and West Volhynian Upland based on inter-simple sequence repeats (ISSR) polymorphism. Twenty ISSR primers amplified 374 DNA fragments, of which 79% were polymorphic. The polymorphic information content values ranged from 0.230 to 0.430 with an average of 0.344. An average genetic similarity calculated based on Dice algorithm between all analysed samples was 0.635 (0.28-1.00). The AMOVA study found a significant difference (Φpt = 0.88, P = 0.001) between Anagallis L. genotypes gathered in Volhynian Polesie (VP) and West Volhynian Upland (VU). Analysis indicated, that 89% of the variation existed among groups and 11% within groups. UPGMA analyses grouped A. foemina samples into 2 clearly separated clusters. The plants of the same geographic origin were grouped together. Principal coordinates analysis (PCoA) as well as STRUCTURE also grouped samples in consistence with the collection site, indicating close genetic affinity of plants from the same location. The observed results are typical for fragmented and isolated populations of rare species. Isolation of a small population leads to a decrease in internal genetic variation and to an increase of variation among them. In that case, the conservation of populations from each regional cluster is important to preserve biodiversity.
Keywords: Genetic similarity; ISSR; Molecular markers; Primulaceae.
© The Author(s) 2020.
Figures




References
-
- Anderberg AA, Ståhl B. Phylogenetic interrelationships in the order Primulales with special emphasis on the family circumscriptions. Can J Bot. 1995;73:1699–1730. doi: 10.1139/b95-184. - DOI
-
- Baessler C, Klotz S. Effects of changes in agricultural land-use on landscape structure and arable weed vegetation over the last 50 years. Agric Ecosyst Environ. 2006;115:43–50. doi: 10.1016/j.agee.2005.12.007. - DOI
-
- Banfi E, Galasso G, Soldano A. Notes on systematics and taxonomy for the Italian vascular flora 1. Atti della Societa italiana di scienze naturalie del Museo civico di storia naturale di Milano. 2005;146:219–244.
-
- Brütting C (2013) Genetic diversity and population structure of arable plants in situ and ex situ—How sustainable is long term cultivation in botanical gardens compared to in situ conditions? Ph.D. dissertation, University Halle-Wittenberg Germany
-
- Brütting C, Meyer S, Kühne P, Hensen I, Wesche K. Spatial genetic structure and low diversity of the rare arable plant Bupleurum rotundifolium L. indicate fragmentation in central Europe. Agric Ecosyst Environ. 2012;161:70–77. doi: 10.1016/j.agee.2012.07.017. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous
