Instability of circular RNAs in clinical tissue samples impairs their reliable expression analysis using RT-qPCR: from the myth of their advantage as biomarkers to reality
- PMID: 32802191
- PMCID: PMC7415809
- DOI: 10.7150/thno.46341
Instability of circular RNAs in clinical tissue samples impairs their reliable expression analysis using RT-qPCR: from the myth of their advantage as biomarkers to reality
Abstract
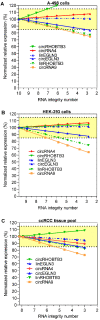
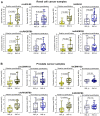
Background: Circular RNAs (circRNAs) are a new class of RNAs with medical significance. Compared to that of linear mRNA transcripts, the stability of circRNAs against degradation owing to their circular structure is considered advantageous for their use as biomarkers. As systematic studies on the stability of circRNAs depending on the RNA integrity, determined as RNA integrity number (RIN), in clinical tissue samples are lacking, we have investigated this aspect in the present study under model and clinical conditions. Methods: Total RNA isolated from kidney cancer tissue and cell lines (A-498 and HEK-293) with different RIN after thermal degradation was used in model experiments. Further, RNA isolated from kidney cancer and prostate cancer tissue collected under routine surgical conditions, representing clinical samples with RIN ranging from 2 to 9, were examined. Quantitative real-time reverse-transcription polymerase chain reaction (RT-qPCR) analysis of several circRNAs (circEGLN3, circRHOBTB3, circCSNK1G3, circRNA4, and circRNA9), their corresponding linear counterparts, tissue-specific reference genes, and three microRNAs (as controls) was performed. The quantification cycles were converted into relative quantities and normalized to the expression of specific reference genes for the corresponding tissue. The effect of RIN on the expression of different RNA entities was determined using linear regression analysis, and clinical samples were classified into two groups based on RIN greater or lesser than 6. Results: The results of model experiments and clinical sample analyses showed that all relative circRNA expression gradually decreased with reduction in RIN values. The adverse effect of RIN was partially compensated after normalizing the data and limiting the samples to only those with RIN values > 6. Conclusions: Our results suggested that circRNAs are not stable in clinical tissue samples, but are subjected to degradative processes similar to mRNAs. This has not been investigated extensively in circRNA expression studies, and hence must be considered in future for obtaining reliable circRNA expression data. This can be achieved by applying the principles commonly used in mRNA expression studies.
Keywords: RNA degradation; RNA integrity; RT-qPCR; circRNA stability; circular RNAs; normalization.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



Similar articles
-
Circular RNAs and Their Linear Transcripts as Diagnostic and Prognostic Tissue Biomarkers in Prostate Cancer after Prostatectomy in Combination with Clinicopathological Factors.Int J Mol Sci. 2020 Oct 22;21(21):7812. doi: 10.3390/ijms21217812. Int J Mol Sci. 2020. PMID: 33105568 Free PMC article.
-
Identification of internal control genes for circular RNAs.Biotechnol Lett. 2019 Oct;41(10):1111-1119. doi: 10.1007/s10529-019-02723-0. Epub 2019 Aug 19. Biotechnol Lett. 2019. PMID: 31428905
-
Robust microRNA stability in degraded RNA preparations from human tissue and cell samples.Clin Chem. 2010 Jun;56(6):998-1006. doi: 10.1373/clinchem.2009.141580. Epub 2010 Apr 8. Clin Chem. 2010. PMID: 20378769
-
A novel strategy of identifying circRNA biomarkers in cardiovascular disease by meta-analysis.J Cell Physiol. 2019 Dec;234(12):21601-21612. doi: 10.1002/jcp.28817. Epub 2019 May 21. J Cell Physiol. 2019. PMID: 31115050 Review.
-
The Use of circRNAs as Biomarkers of Cancer.Methods Mol Biol. 2021;2348:307-341. doi: 10.1007/978-1-0716-1581-2_21. Methods Mol Biol. 2021. PMID: 34160816 Review.
Cited by
-
Cost-Effective Transcriptome-Wide Profiling of Circular RNAs by the Improved-tdMDA-NGS Method.Front Mol Biosci. 2022 May 13;9:886366. doi: 10.3389/fmolb.2022.886366. eCollection 2022. Front Mol Biosci. 2022. PMID: 35647023 Free PMC article.
-
Multiple roles of circular RNAs in prostate cancer: from the biological basis to potential clinical applications.Eur J Med Res. 2025 Feb 27;30(1):140. doi: 10.1186/s40001-025-02382-0. Eur J Med Res. 2025. PMID: 40016786 Free PMC article. Review.
-
Exploratory Circular RNA Profiling in Adrenocortical Tumors.Cancers (Basel). 2022 Sep 2;14(17):4313. doi: 10.3390/cancers14174313. Cancers (Basel). 2022. PMID: 36077848 Free PMC article.
-
Circular RNAs in peripheral blood mononuclear cells are more stable than linear RNAs upon sample processing delay.J Cell Mol Med. 2022 Oct;26(19):5021-5032. doi: 10.1111/jcmm.17525. Epub 2022 Aug 30. J Cell Mol Med. 2022. PMID: 36039821 Free PMC article.
-
CRISPR/Cas13-Based Platforms for a Potential Next-Generation Diagnosis of Colorectal Cancer through Exosomes Micro-RNA Detection: A Review.Cancers (Basel). 2021 Sep 16;13(18):4640. doi: 10.3390/cancers13184640. Cancers (Basel). 2021. PMID: 34572866 Free PMC article. Review.
References
-
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8. - PubMed
-
- Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S. et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–85. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

