Computational advances of tumor marker selection and sample classification in cancer proteomics
- PMID: 32802273
- PMCID: PMC7403885
- DOI: 10.1016/j.csbj.2020.07.009
Computational advances of tumor marker selection and sample classification in cancer proteomics
Abstract
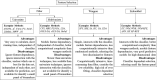
Cancer proteomics has become a powerful technique for characterizing the protein markers driving transformation of malignancy, tracing proteome variation triggered by therapeutics, and discovering the novel targets and drugs for the treatment of oncologic diseases. To facilitate cancer diagnosis/prognosis and accelerate drug target discovery, a variety of methods for tumor marker identification and sample classification have been developed and successfully applied to cancer proteomic studies. This review article describes the most recent advances in those various approaches together with their current applications in cancer-related studies. Firstly, a number of popular feature selection methods are overviewed with objective evaluation on their advantages and disadvantages. Secondly, these methods are grouped into three major classes based on their underlying algorithms. Finally, a variety of sample separation algorithms are discussed. This review provides a comprehensive overview of the advances on tumor maker identification and patients/samples/tissues separations, which could be guidance to the researches in cancer proteomics.
Keywords: ANN, Artificial Neural Network; ANOVA, Analysis of Variance; CFS, Correlation-based Feature Selection; Cancer proteomics; Computational methods; DAPC, Discriminant Analysis of Principal Component; DT, Decision Trees; EDA, Estimation of Distribution Algorithm; FC, Fold Change; GA, Genetic Algorithms; GR, Gain Ratio; HC, Hill Climbing; HCA, Hierarchical Cluster Analysis; IG, Information Gain; LDA, Linear Discriminant Analysis; LIMMA, Linear Models for Microarray Data; MBF, Markov Blanket Filter; MWW, Mann–Whitney–Wilcoxon test; OPLS-DA, Orthogonal Partial Least Squares Discriminant Analysis; PCA, Principal Component Analysis; PLS-DA, Partial Least Square Discriminant Analysis; RF, Random Forest; RF-RFE, Random Forest with Recursive Feature Elimination; SA, Simulated Annealing; SAM, Significance Analysis of Microarrays; SBE, Sequential Backward Elimination; SFS, and Sequential Forward Selection; SOM, Self-organizing Map; SU, Symmetrical Uncertainty; SVM, Support Vector Machine; SVM-RFE, Support Vector Machine with Recursive Feature Elimination; Sample classification; Tumor marker selection; sPLSDA, Sparse Partial Least Squares Discriminant Analysis; t-SNE, Student t Distribution; χ2, Chi-square.
© 2020 The Authors.
Figures
References
-
- Malvezzi M., Carioli G., Bertuccio P., Negri E., La Vecchia C. Relation between mortality trends of cardiovascular diseases and selected cancers in the European Union, in 1970–2017. Focus on cohort and period effects. Eur J Cancer. 2018;103:341–355. - PubMed
-
- Arora D., Chaudhary R., Singh A. System biology approach to identify potential receptor for targeting cancer and biomolecular interaction studies of indole[2,1-a]isoquinoline derivative as anticancerous drug candidate against it. Interdiscip Sci Comput Life Sci. 2017;11:125–134. - PubMed
Publication types
LinkOut - more resources
Full Text Sources