Potential chimeric peptides to block the SARS-CoV-2 spike receptor-binding domain
- PMID: 32802318
- PMCID: PMC7411520
- DOI: 10.12688/f1000research.24074.1
Potential chimeric peptides to block the SARS-CoV-2 spike receptor-binding domain
Abstract
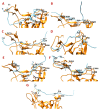
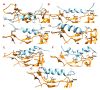
Background: There are no known medicines or vaccines to control the COVID-19 pandemic caused by SARS-CoV-2 (nCoV). Antiviral peptides are superior to conventional drugs and may also be effective against COVID-19. Hence, we investigated the SARS-CoV-2 Spike receptor-binding domain (nCoV-RBD) that interacts with hACE2 for viral attachment and entry. Methods: Three strategies and bioinformatics approaches were employed to design potential nCoV-RBD - hACE2 interaction-blocking peptides that may restrict viral attachment and entry. Firstly, the key residues interacting with nCoV-RBD - hACE2 are identified and hACE2 sequence-based peptides are designed. Second, peptides from five antibacterial peptide databases that block nCoV-RBD are identified; finally, a chimeric peptide design approach is used to design peptides that can bind to key nCoV-RBD residues. The final peptides are selected based on their physiochemical properties, numbers and positions of key residues binding, binding energy, and antiviral properties. Results: We found that: (i) three amino acid stretches in hACE2 interact with nCoV-RBD; (ii) effective peptides must bind to three key positions of nCoV-RBD (Gly485/Phe486/Asn487, Gln493, and Gln498/Thr500/Asn501); (iii) Phe486, Gln493, and Asn501 are critical residues; (iv) AC20 and AC23 derived from hACE2 may block two key critical positions; (iv) DBP6 identified from databases can block the three sites of the nCoV-RBD and interacts with one critical position, Gln498; (v) seven chimeric peptides were considered promising, among which cnCoVP-3, cnCoVP-4, and cnCoVP-7 are the top three; and (vi) cnCoVP-4 meets all the criteria and is the best peptide. Conclusions: To conclude, using three different bioinformatics approaches, we identified 17 peptides that can potentially bind to the nCoV-RBD that interacts with hACE2. Binding these peptides to nCoV-RBD may potentially inhibit the virus to access hACE2 and thereby may prevent the infection. Out of 17, 10 peptides have promising potential and need further experimental validation.
Keywords: ACE2; Antiviral peptides; COVID-19; SARS-CoV-2; Spike protein; nCoV-19; peptide design.
Copyright: © 2020 Barh D et al.
Conflict of interest statement
No competing interests were disclosed.
Figures
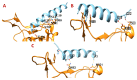
References
-
- Singh AK, Singh A, Shaikh A, et al. : Chloroquine and hydroxychloroquine in the treatment of covid-19 with or without diabetes: A systematic search and a narrative review with a special reference to india and other developing countries. Diabetes Metab Syndr. 2020;14(3):241–246. 10.1016/j.dsx.2020.03.011 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

