ITGA4 gene methylation status in chronic lymphocytic leukemia
- PMID: 32802392
- PMCID: PMC7421236
- DOI: 10.2144/fsoa-2020-0034
ITGA4 gene methylation status in chronic lymphocytic leukemia
Abstract
Background: We aimed to investigate ITGA4 gene expression pattern and to explore its methylation heterogeneity in chronic lymphocytic leukemia (CLL).
Patients & methods: Eighty one CLL patients and 75 healthy subjects were enrolled and prognostic evaluation of patients was assessed. ITGA4 q-realtime PCR was performed using Applied Biosystems, TaqMan gene expression assay. ITGA4 gene-specific CpG methylation was investigated in real time using pyrosequencing technology.
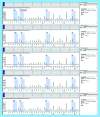
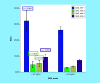
Results: ITGA4 was differentially expressed in CLL patients. The CpG sites-1, 2 and 3 showed significantly higher mean levels than healthy controls (p = <0.001, 0.007 and 0.009). Significant association between CpG site-1 and CLL has been detected using age-adjusted logistic regression (p < 0.001).
Conclusion: Hypermethylation at ITGA4 gene CpG sites (1,2,3) is a characteristic feature in CLL.
Keywords: ITGA4 gene expression; beta 2 microglobulin; chronic lymphocytic leukemia; cytogenetics; methylation analysis; prognostic markers; pyrosequencing.
© 2020 Hanaa RM Attia.
Conflict of interest statement
Financial & competing interests disclosure The authors gratefully acknowledge the financial support of the Science and Technology Development Fund (STDF), Egypt through Targeted Health call grant no. 22918 and Capacity Building Grant no. 4880. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript.
Figures



References
-
- Stilgenbauer S, Schnaiter A, Paschka P. et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood 123(21), 3247–3254 (2014). - PubMed
-
- Rassenti LZ, Huynh L, Toy TL. et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N. Engl. J. Med. 351(9), 893–901 (2004). - PubMed
-
- Bulian P, Shanafelt TD, Fegan C. et al. CD49d is the strongest flow cytometry-based predictor of overall survival in chronic lymphocytic leukemia. J. Clin. Oncol. 32(9), 897–904 (2014). - PMC - PubMed
-
• In this analysis of approximately 3000 patients with chronic lymphocytic leukemia (CLL), authors identified CD49d as the strongest flow cytometry-based predictor of overall survival (OS) and treatment-free survival (TFS) independent of CD38 and ZAP-70.
-
- Zucchetto A, Caldana C, Benedetti D. et al. CD49d is overexpressed by trisomy 12 chronic lymphocytic leukemia cells: evidence for methylation-dependent regulation mechanism. Blood 122(19), 3317–3321 (2013). - PubMed
-
• Authors demonstrated that CD49 is universally expressed in Trisomy12 CLL and is regulated by a methylation-dependent mechanism
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
