This is a preprint.
Identification of SARS-CoV-2 3CL Protease Inhibitors by a Quantitative High-throughput Screening
- PMID: 32803196
- PMCID: PMC7427131
- DOI: 10.1101/2020.07.17.207019
Identification of SARS-CoV-2 3CL Protease Inhibitors by a Quantitative High-throughput Screening
Update in
-
Identification of SARS-CoV-2 3CL Protease Inhibitors by a Quantitative High-Throughput Screening.ACS Pharmacol Transl Sci. 2020 Sep 4;3(5):1008-1016. doi: 10.1021/acsptsci.0c00108. eCollection 2020 Oct 9. ACS Pharmacol Transl Sci. 2020. PMID: 33062953 Free PMC article.
Abstract
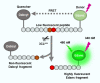
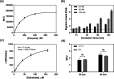
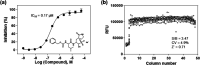
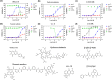
The outbreak of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has emphasized the urgency to develop effective therapeutics. Drug repurposing screening is regarded as one of the most practical and rapid approaches for the discovery of such therapeutics. The 3C like protease (3CL pro ), or main protease (M pro ) of SARS-CoV-2 is a valid drug target as it is a specific viral enzyme and plays an essential role in viral replication. We performed a quantitative high throughput screening (qHTS) of 10,755 compounds consisting of approved and investigational drugs, and bioactive compounds using a SARS-CoV-2 3CL pro assay. Twenty-three small molecule inhibitors of SARS-CoV-2 3CL pro have been identified with IC50s ranging from 0.26 to 28.85 μM. Walrycin B (IC 50 = 0.26 µM), Hydroxocobalamin (IC 50 = 3.29 µM), Suramin sodium (IC 50 = 6.5 µM), Z-DEVD-FMK (IC 50 = 6.81 µM), LLL-12 (IC 50 = 9.84 µM), and Z-FA-FMK (IC 50 = 11.39 µM) are the most potent 3CL pro inhibitors. The activities of anti-SARS-CoV-2 viral infection was confirmed in 7 of 23 compounds using a SARS-CoV-2 cytopathic effect assay. The results demonstrated a set of SARS-CoV-2 3CL pro inhibitors that may have potential for further clinical evaluation as part of drug combination therapies to treating COVID-19 patients, and as starting points for chemistry optimization for new drug development.
Conflict of interest statement
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Figures


References
-
- Wu F.; Zhao S.; Yu B.; Chen Y.-M.; Wang W.; Song Z.-G.; Hu Y.; Tao Z.-W.; Tian J.-H.; Pei Y.-Y.; Yuan M.-L.; Zhang Y.-L.; Dai F.-H.; Liu Y.; Wang Q.-M.; Zheng J.-J.; Xu L.; Holmes E. C.; Zhang Y.-Z., A new coronavirus associated with human respiratory disease in China. Nature 2020, 579 (7798), 265–269. - PMC - PubMed
-
- Jin Z.; Du X.; Xu Y.; Deng Y.; Liu M.; Zhao Y.; Zhang B.; Li X.; Zhang L.; Peng C.; Duan Y.; Yu J.; Wang L.; Yang K.; Liu F.; Jiang R.; Yang X.; You T.; Liu X.; Yang X.; Bai F.; Liu H.; Liu X.; Guddat L. W.; Xu W.; Xiao G.; Qin C.; Shi Z.; Jiang H.; Rao Z.; Yang H., Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582 (7811), 289–293. - PubMed
-
- Thiel V.; Ivanov K. A.; Putics Á.; Hertzig T.; Schelle B.; Bayer S.; Weißbrich B.; Snijder E. J.; Rabenau H.; Doerr H. W.; Gorbalenya A. E.; Ziebuhr J., Mechanisms and enzymes involved in SARS coronavirus genome expression. Journal of General Virology 2003, 84 (9), 2305–2315. - PubMed
-
- Anand K.; Ziebuhr J Fau - Wadhwani P.; Wadhwani P Fau - Mesters J. R.; Mesters Fau Jr - Hilgenfeld R.; Hilgenfeld R., Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 2003, 300 (5626), 1763–1767. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Chemical Information
Miscellaneous
