This is a preprint.
Cryo-EM Structures of the SARS-CoV-2 Endoribonuclease Nsp15
- PMID: 32803198
- PMCID: PMC7427136
- DOI: 10.1101/2020.08.11.244863
Cryo-EM Structures of the SARS-CoV-2 Endoribonuclease Nsp15
Update in
-
Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics.Nat Commun. 2021 Jan 27;12(1):636. doi: 10.1038/s41467-020-20608-z. Nat Commun. 2021. PMID: 33504779 Free PMC article.
Abstract
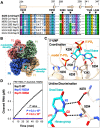
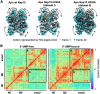
New therapeutics are urgently needed to inhibit SARS-CoV-2, the virus responsible for the on-going Covid-19 pandemic. Nsp15, a uridine-specific endoribonuclease found in all coronaviruses, processes viral RNA to evade detection by RNA-activated host defense systems, making it a promising drug target. Previous work with SARS-CoV-1 established that Nsp15 is active as a hexamer, yet how Nsp15 recognizes and processes viral RNA remains unknown. Here we report a series of cryo-EM reconstructions of SARS-CoV-2 Nsp15. The UTP-bound cryo-EM reconstruction at 3.36 Å resolution provides molecular details into how critical residues within the Nsp15 active site recognize uridine and facilitate catalysis of the phosphodiester bond, whereas the apo-states reveal active site conformational heterogeneity. We further demonstrate the specificity and mechanism of nuclease activity by analyzing Nsp15 products using mass spectrometry. Collectively, these findings advance understanding of how Nsp15 processes viral RNA and provide a structural framework for the development of new therapeutics.
Conflict of interest statement
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
