Genome-wide Identification of WRKY transcription factor family members in sorghum (Sorghum bicolor (L.) moench)
- PMID: 32804948
- PMCID: PMC7430707
- DOI: 10.1371/journal.pone.0236651
Genome-wide Identification of WRKY transcription factor family members in sorghum (Sorghum bicolor (L.) moench)
Abstract
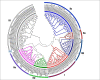
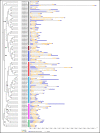
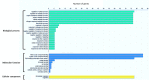
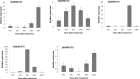
WRKY transcription factors regulate diverse biological processes in plants, including abiotic and biotic stress responses, and constitute one of the largest transcription factor families in higher plants. Although the past decade has seen significant progress towards identifying and functionally characterizing WRKY genes in diverse species, little is known about the WRKY family in sorghum (Sorghum bicolor (L.) moench). Here we report the comprehensive identification of 94 putative WRKY transcription factors (SbWRKYs). The SbWRKYs were divided into three groups (I, II, and III), with those in group II further classified into five subgroups (IIa-IIe), based on their conserved domains and zinc finger motif types. WRKYs from the model plant Arabidopsis (Arabidopsis thaliana) were used for the phylogenetic analysis of all SbWRKY genes. Motif analysis showed that all SbWRKYs contained either one or two WRKY domains and that SbWRKYs within the same group had similar motif compositions. SbWRKY genes were located on all 10 sorghum chromosomes, and some gene clusters and two tandem duplications were detected. SbWRKY gene structure analysis showed that they contained 0-7 introns, with most SbWRKY genes consisting of two introns and three exons. Gene ontology (GO) annotation functionally categorized SbWRKYs under cellular components, molecular functions and biological processes. A cis-element analysis showed that all SbWRKYs contain at least one stress response-related cis-element. We exploited publicly available microarray datasets to analyze the expression profiles of 78 SbWRKY genes at different growth stages and in different tissues. The induction of SbWRKYs by different abiotic stresses hinted at their potential involvement in stress responses. qRT-PCR analysis revealed different expression patterns for SbWRKYs during drought stress. Functionally characterized WRKY genes in Arabidopsis and other species will provide clues for the functional characterization of putative orthologs in sorghum. Thus, the present study delivers a solid foundation for future functional studies of SbWRKY genes and their roles in the response to critical stresses such as drought.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

