Bacterial biofilms colonizing plastics in estuarine waters, with an emphasis on Vibrio spp. and their antibacterial resistance
- PMID: 32804963
- PMCID: PMC7430737
- DOI: 10.1371/journal.pone.0237704
Bacterial biofilms colonizing plastics in estuarine waters, with an emphasis on Vibrio spp. and their antibacterial resistance
Abstract
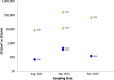
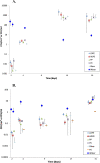
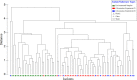
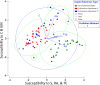
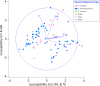
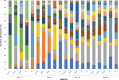
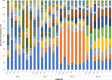
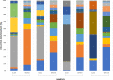
Since plastics degrade very slowly, they remain in the environment on much longer timescales than most natural organic substrates and provide a novel habitat for colonization by bacterial communities. The spectrum of relationships between plastics and bacteria, however, is little understood. The first objective of this study was to examine plastics as substrates for communities of Bacteria in estuarine surface waters. We used next-generation sequencing of the 16S rRNA gene to characterize communities from plastics collected in the field, and over the course of two colonization experiments, from biofilms that developed on plastic (low-density polyethylene, high-density polyethylene, polypropylene, polycarbonate, polystyrene) and glass substrates placed in the environment. Both field sampling and colonization experiments were conducted in estuarine tributaries of the lower Chesapeake Bay. As a second objective, we concomitantly analyzed biofilms on plastic substrates to ascertain the presence and abundance of Vibrio spp. bacteria, then isolated three human pathogens, V. cholerae, V. parahaemolyticus, and V. vulnificus, and determined their antibiotic-resistant profiles. In both components of this study, we compared our results with analyses conducted on paired samples of estuarine water. This research adds to a nascent literature that suggests environmental factors govern the development of bacterial communities on plastics, more so than the characteristics of the plastic substrates themselves. In addition, this study is the first to culture three pathogenic vibrios from plastics in estuaries, reinforcing and expanding upon earlier reports of plastic pollution as a habitat for Vibrio species. The antibiotic resistance detected among the isolates, coupled with the longevity of plastics in the aqueous environment, suggests biofilms on plastics have potential to persist and serve as focal points of potential pathogens and horizontal gene transfer.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Obbard RW, Sadri S, Wong YQ, Khitun AA, Baker I, Thompson RC. Global warming releases microplastic legacy frozen in Arctic Sea ice. Earth’s Future 2: 315–320.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

