Development of novel g-SSR markers in guava (Psidium guajava L.) cv. Allahabad Safeda and their application in genetic diversity, population structure and cross species transferability studies
- PMID: 32804981
- PMCID: PMC7431106
- DOI: 10.1371/journal.pone.0237538
Development of novel g-SSR markers in guava (Psidium guajava L.) cv. Allahabad Safeda and their application in genetic diversity, population structure and cross species transferability studies
Abstract
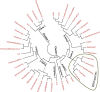
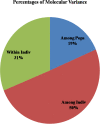
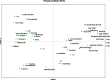
Dearth of genomic resources particularly, microsatellite markers in nutritionally and commercially important fruit crop, guava necessitate the development of the novel genomic SSR markers through the library enrichment techniques. Three types of 3' -biotinylated oligonucleotide probes [(CT)14, (GT)12, and (AAC)8] were used to develop microsatellite enriched libraries. A total of 153 transformed colonies were screened of which 111 positive colonies were subjected for Sanger sequencing. The clones having more than five motif repeats were selected for primer designing and a total of 38 novel genomic simple sequence repeats could be identified. The g-SSRs had the motif groups ranging from monomer to pentamer out of which dimer group occurred the most (89.47%). Out of 38 g-SSRs markers developed, 26 were found polymorphic, which showed substantial genetic diversity among the guava genotypes including wild species. The average number of alleles per locus, major allele frequency, gene diversity, expected heterozygosity and polymorphic information content of 26 SSRs were 3.46, 0.56, 0.53, 0.29 and 0.46, respectively. The rate of cross-species transferability of the developed g-SSR loci varied from 38.46 to 80.77% among the studied wild Psidium species. Generation of N-J tree based on 26 SSRs grouped the 40 guava genotypes into six clades with two out-groups, the wild guava species showed genetic distinctness from cultivated genotypes. Furthermore, population structure analysis grouped the guava genotypes into three genetic groups, which were partly supported by PCoA and N-J tree. Further, AMOVA and PCoA deciphered high genetic diversity among the present set of guava genotypes including wild species. Thus, the developed novel g-SSRs were found efficient and informative for diversity and population structure analyses of the guava genotypes. These developed novel g-SSR loci would add to the new genomic resource in guava, which may be utilized in genomic-assisted guava breeding.
Conflict of interest statement
The authors have declared no competing interests exist.
Figures



Similar articles
-
Development of genome-wide SSR markers through in silico mining of guava (Psidium guajava L.) genome for genetic diversity analysis and transferability studies across species and genera.Front Plant Sci. 2025 Apr 25;16:1527866. doi: 10.3389/fpls.2025.1527866. eCollection 2025. Front Plant Sci. 2025. PMID: 40353228 Free PMC article.
-
Transferability of simple sequence repeat (SSR) markers developed in guava (Psidium guajava L.) to four Myrtaceae species.Mol Biol Rep. 2013 Aug;40(8):5067-71. doi: 10.1007/s11033-013-2608-1. Epub 2013 May 9. Mol Biol Rep. 2013. PMID: 23657599
-
Development of Genome-Wide Functional Markers Using Draft Genome Assembly of Guava (Psidium guajava L.) cv. Allahabad Safeda to Expedite Molecular Breeding.Front Plant Sci. 2021 Sep 23;12:708332. doi: 10.3389/fpls.2021.708332. eCollection 2021. Front Plant Sci. 2021. PMID: 34630458 Free PMC article.
-
RNA-sequencing based gene expression landscape of guava cv. Allahabad Safeda and comparative analysis to colored cultivars.BMC Genomics. 2020 Jul 15;21(1):484. doi: 10.1186/s12864-020-06883-6. BMC Genomics. 2020. PMID: 32669108 Free PMC article.
-
Streamlining of Simple Sequence Repeat Data Mining Methodologies and Pipelines for Crop Scanning.Plants (Basel). 2024 Sep 19;13(18):2619. doi: 10.3390/plants13182619. Plants (Basel). 2024. PMID: 39339594 Free PMC article. Review.
Cited by
-
Construction of Genetic Linkage Map and Mapping QTL Specific to Leaf Anthocyanin Colouration in Mapping Population 'Allahabad Safeda' × 'Purple Guava (Local)' of Guava (Psidium guajava L.).Plants (Basel). 2022 Aug 2;11(15):2014. doi: 10.3390/plants11152014. Plants (Basel). 2022. PMID: 35956491 Free PMC article.
-
Construction of a genetic linkage map and QTL mapping of fruit quality traits in guava (Psidium guajava L.).Front Plant Sci. 2023 Jun 22;14:1123274. doi: 10.3389/fpls.2023.1123274. eCollection 2023. Front Plant Sci. 2023. PMID: 37426984 Free PMC article.
-
Assessing Genetic Variation among Strychnos spinosa Lam. Morphotypes Using Simple Sequence Repeat Markers.Plants (Basel). 2023 Jul 28;12(15):2810. doi: 10.3390/plants12152810. Plants (Basel). 2023. PMID: 37570964 Free PMC article.
-
Development of genome-wide SSR markers through in silico mining of guava (Psidium guajava L.) genome for genetic diversity analysis and transferability studies across species and genera.Front Plant Sci. 2025 Apr 25;16:1527866. doi: 10.3389/fpls.2025.1527866. eCollection 2025. Front Plant Sci. 2025. PMID: 40353228 Free PMC article.
-
DNA Barcode, chemical analysis, and antioxidant activity of Psidium guineense from Ecuador.PLoS One. 2025 Mar 19;20(3):e0319524. doi: 10.1371/journal.pone.0319524. eCollection 2025. PLoS One. 2025. PMID: 40106513 Free PMC article.
References
-
- Nakasone HY, Paull RE. Tropical fruits. Cab International; 1998.
-
- Samson JA. Tropical fruits-Tropical agricultural series. New York: Longman Inc; 1984.
-
- Morton JF, Dowling CF. Fruits of warm climates. Miami, FL: JF Morton; 1987.
-
- Rajan S, Hudedamani U. Genetic Resources of Guava: Importance, Uses and Prospects In: Rajasekharan PE, Rao VR, editors. Conservation and Utilization of Horticultural Genetic Resources. Singapore: Springer; 2019. p.363–383.
-
- NHB. National Horticulture Database. National Horticulture Board, Gurgram, Haryana; 2018. 10.1093/database/bay088 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

