Structures and distributions of SARS-CoV-2 spike proteins on intact virions
- PMID: 32805734
- PMCID: PMC7116492
- DOI: 10.1038/s41586-020-2665-2
Structures and distributions of SARS-CoV-2 spike proteins on intact virions
Abstract
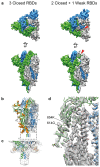
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virions are surrounded by a lipid bilayer from which spike (S) protein trimers protrude1. Heavily glycosylated S trimers bind to the angiotensin-converting enzyme 2 receptor and mediate entry of virions into target cells2-6. S exhibits extensive conformational flexibility: it modulates exposure of its receptor-binding site and subsequently undergoes complete structural rearrangement to drive fusion of viral and cellular membranes2,7,8. The structures and conformations of soluble, overexpressed, purified S proteins have been studied in detail using cryo-electron microscopy2,7,9-12, but the structure and distribution of S on the virion surface remain unknown. Here we applied cryo-electron microscopy and tomography to image intact SARS-CoV-2 virions and determine the high-resolution structure, conformational flexibility and distribution of S trimers in situ on the virion surface. These results reveal the conformations of S on the virion, and provide a basis from which to understand interactions between S and neutralizing antibodies during infection or vaccination.
Conflict of interest statement
Figures
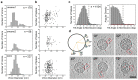

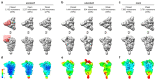


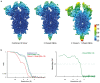
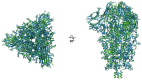


Comment in
-
SARS-CoV-2 spike behavior in situ: a Cryo-EM images for a better understanding of the COVID-19 pandemic.Signal Transduct Target Ther. 2020 Oct 30;5(1):252. doi: 10.1038/s41392-020-00365-7. Signal Transduct Target Ther. 2020. PMID: 33127886 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

