Dynamic nanopore long-read sequencing analysis of HIV-1 splicing events during the early steps of infection
- PMID: 32807178
- PMCID: PMC7433067
- DOI: 10.1186/s12977-020-00533-1
Dynamic nanopore long-read sequencing analysis of HIV-1 splicing events during the early steps of infection
Abstract
Background: Alternative splicing is a key step in Human Immunodeficiency Virus type 1 (HIV-1) replication that is tightly regulated both temporally and spatially. More than 50 different transcripts can be generated from a single HIV-1 unspliced pre-messenger RNA (pre-mRNA) and a balanced proportion of unspliced and spliced transcripts is critical for the production of infectious virions. Understanding the mechanisms involved in the regulation of viral RNA is therefore of potential therapeutic interest. However, monitoring the regulation of alternative splicing events at a transcriptome-wide level during cell infection is challenging. Here we used the long-read cDNA sequencing developed by Oxford Nanopore Technologies (ONT) to explore in a quantitative manner the complexity of the HIV-1 transcriptome regulation in infected primary CD4+ T cells.
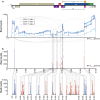
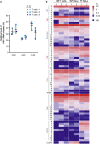
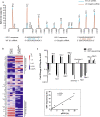
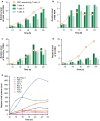
Results: ONT reads mapping to the viral genome proved sufficiently long to span all possible splice junctions, even distant ones, and to be assigned to a total of 150 exon combinations. Fifty-three viral RNA isoforms, including 14 new ones were further considered for quantification. Relative levels of viral RNAs determined by ONT sequencing showed a high degree of reproducibility, compared favourably to those produced in previous reports and highly correlated with quantitative PCR (qPCR) data. To get further insights into alternative splicing regulation, we then compiled quantifications of splice site (SS) usage and transcript levels to build "splice trees", a quantitative representation of the cascade of events leading to the different viral isoforms. This approach allowed visualizing the complete rewiring of SS usages upon perturbation of SS D2 and its impact on viral isoform levels. Furthermore, we produced the first dynamic picture of the cascade of events occurring between 12 and 24 h of viral infection. In particular, our data highlighted the importance of non-coding exons in viral RNA transcriptome regulation.
Conclusion: ONT sequencing is a convenient and reliable strategy that enabled us to grasp the dynamic of the early splicing events modulating the viral RNA landscape in HIV-1 infected cells.
Keywords: Alternative splicing; HIV RNA; ONT long-read sequencing; Viral transcriptome.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- De Conti L, Baralle M, Buratti E. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip Rev RNA. 2013;4:49–60. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

