HIF-1α and HIF-2α differently regulate tumour development and inflammation of clear cell renal cell carcinoma in mice
- PMID: 32807776
- PMCID: PMC7431415
- DOI: 10.1038/s41467-020-17873-3
HIF-1α and HIF-2α differently regulate tumour development and inflammation of clear cell renal cell carcinoma in mice
Abstract
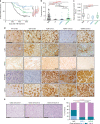
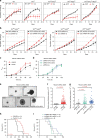
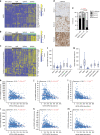
Mutational inactivation of VHL is the earliest genetic event in the majority of clear cell renal cell carcinomas (ccRCC), leading to accumulation of the HIF-1α and HIF-2α transcription factors. While correlative studies of human ccRCC and functional studies using human ccRCC cell lines have implicated HIF-1α as an inhibitor and HIF-2α as a promoter of aggressive tumour behaviours, their roles in tumour onset have not been functionally addressed. Herein we show using an autochthonous ccRCC model that Hif1a is essential for tumour formation whereas Hif2a deletion has only minor effects on tumour initiation and growth. Both HIF-1α and HIF-2α are required for the clear cell phenotype. Transcriptomic and proteomic analyses reveal that HIF-1α regulates glycolysis while HIF-2α regulates genes associated with lipoprotein metabolism, ribosome biogenesis and E2F and MYC transcriptional activities. HIF-2α-deficient tumours are characterised by increased antigen presentation, interferon signalling and CD8+ T cell infiltration and activation. Single copy loss of HIF1A or high levels of HIF2A mRNA expression correlate with altered immune microenvironments in human ccRCC. These studies reveal an oncogenic role of HIF-1α in ccRCC initiation and suggest that alterations in the balance of HIF-1α and HIF-2α activities can affect different aspects of ccRCC biology and disease aggressiveness.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures




References
-
- Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. - PubMed
-
- Frew IJ, Moch H. A clearer view of the molecular complexity of clear cell renal cell carcinoma. Annu. Rev. Pathol. Mech. Dis. 2015;10:263–289. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

