CTCF-mediated chromatin looping in EGR2 regulation and SUZ12 recruitment critical for peripheral myelination and repair
- PMID: 32807777
- PMCID: PMC7431862
- DOI: 10.1038/s41467-020-17955-2
CTCF-mediated chromatin looping in EGR2 regulation and SUZ12 recruitment critical for peripheral myelination and repair
Abstract
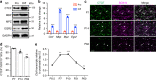
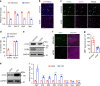
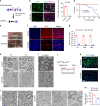
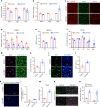
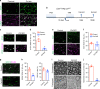
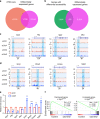
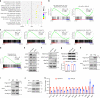
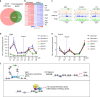
Chromatin organization is critical for cell growth, differentiation, and disease development, however, its functions in peripheral myelination and myelin repair remain elusive. In this report, we demonstrate that the CCCTC-binding factor (CTCF), a crucial chromatin organizer, is essential for Schwann cell myelination and myelin regeneration after nerve injury. Inhibition of CTCF or its deletion blocks Schwann cell differentiation at the pro-myelinating stage, whereas overexpression of CTCF promotes the myelination program. We find that CTCF establishes chromatin interaction loops between enhancer and promoter regulatory elements and promotes expression of a key pro-myelinogenic factor EGR2. In addition, CTCF interacts with SUZ12, a component of polycomb-repressive-complex 2 (PRC2), to repress the transcriptional program associated with negative regulation of Schwann cell maturation. Together, our findings reveal a dual role of CTCF-dependent chromatin organization in promoting myelinogenic programs and recruiting chromatin-repressive complexes to block Schwann cell differentiation inhibitors to control peripheral myelination and repair.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Dynamic regulation of Schwann cell enhancers after peripheral nerve injury.J Biol Chem. 2015 Mar 13;290(11):6937-50. doi: 10.1074/jbc.M114.622878. Epub 2015 Jan 22. J Biol Chem. 2015. PMID: 25614629 Free PMC article.
-
Regulation of Peripheral Nerve Myelin Maintenance by Gene Repression through Polycomb Repressive Complex 2.J Neurosci. 2015 Jun 3;35(22):8640-52. doi: 10.1523/JNEUROSCI.2257-14.2015. J Neurosci. 2015. PMID: 26041929 Free PMC article.
-
Locus-wide identification of Egr2/Krox20 regulatory targets in myelin genes.J Neurochem. 2010 Dec;115(6):1409-20. doi: 10.1111/j.1471-4159.2010.07045.x. Epub 2010 Nov 4. J Neurochem. 2010. PMID: 21044070 Free PMC article.
-
Regulation of myelin-specific gene expression. Relevance to CMT1.Ann N Y Acad Sci. 1999 Sep 14;883:91-108. Ann N Y Acad Sci. 1999. PMID: 10586235 Review.
-
The structural and functional roles of CTCF in the regulation of cell type-specific and human disease-associated super-enhancers.Genes Genomics. 2019 Mar;41(3):257-265. doi: 10.1007/s13258-018-0768-z. Epub 2018 Nov 19. Genes Genomics. 2019. PMID: 30456521 Review.
Cited by
-
Modulation of the high-order chromatin structure by Polycomb complexes.Front Cell Dev Biol. 2022 Oct 5;10:1021658. doi: 10.3389/fcell.2022.1021658. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36274840 Free PMC article. Review.
-
Modulation of Schwann cell homeostasis by the BAP1 deubiquitinase.Glia. 2023 Jun;71(6):1466-1480. doi: 10.1002/glia.24351. Epub 2023 Feb 15. Glia. 2023. PMID: 36790040 Free PMC article.
-
Single-cell multi-omics sequencing uncovers region-specific plasticity of glioblastoma for complementary therapeutic targeting.Sci Adv. 2024 Nov 22;10(47):eadn4306. doi: 10.1126/sciadv.adn4306. Epub 2024 Nov 22. Sci Adv. 2024. PMID: 39576855 Free PMC article.
-
An oligodendrocyte silencer element underlies the pathogenic impact of lamin B1 structural variants.Nat Commun. 2025 Feb 5;16(1):1373. doi: 10.1038/s41467-025-56378-9. Nat Commun. 2025. PMID: 39910058 Free PMC article.
-
Schwann cells acquire a repair phenotype after assembling into spheroids and show enhanced in vivo therapeutic potential for promoting peripheral nerve repair.Bioeng Transl Med. 2023 Dec 26;9(2):e10635. doi: 10.1002/btm2.10635. eCollection 2024 Mar. Bioeng Transl Med. 2023. PMID: 38435829 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

