Identifying proteins bound to native mitotic ESC chromosomes reveals chromatin repressors are important for compaction
- PMID: 32807789
- PMCID: PMC7431861
- DOI: 10.1038/s41467-020-17823-z
Identifying proteins bound to native mitotic ESC chromosomes reveals chromatin repressors are important for compaction
Abstract
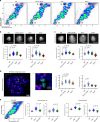
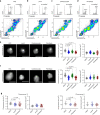
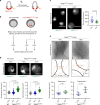
Epigenetic information is transmitted from mother to daughter cells through mitosis. Here, to identify factors that might play a role in conveying epigenetic memory through cell division, we report on the isolation of unfixed, native chromosomes from metaphase-arrested cells using flow cytometry and perform LC-MS/MS to identify chromosome-bound proteins. A quantitative proteomic comparison between metaphase-arrested cell lysates and chromosome-sorted samples reveals a cohort of proteins that were significantly enriched on mitotic ESC chromosomes. These include pluripotency-associated transcription factors, repressive chromatin-modifiers such as PRC2 and DNA methyl-transferases, and proteins governing chromosome architecture. Deletion of PRC2, Dnmt1/3a/3b or Mecp2 in ESCs leads to an increase in the size of individual mitotic chromosomes, consistent with de-condensation. Similar results were obtained by the experimental cleavage of cohesin. Thus, we identify chromosome-bound factors in pluripotent stem cells during mitosis and reveal that PRC2, DNA methylation and Mecp2 are required to maintain chromosome compaction.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 2004;279:48350–48359. - PubMed
-
- Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 2009;10:192–206. - PubMed
-
- Petryk N, et al. MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science. 2018;361:1389–1392. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

