Deployable CRISPR-Cas13a diagnostic tools to detect and report Ebola and Lassa virus cases in real-time
- PMID: 32807807
- PMCID: PMC7431545
- DOI: 10.1038/s41467-020-17994-9
Deployable CRISPR-Cas13a diagnostic tools to detect and report Ebola and Lassa virus cases in real-time
Abstract
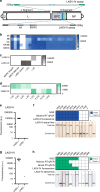
Recent outbreaks of viral hemorrhagic fevers (VHFs), including Ebola virus disease (EVD) and Lassa fever (LF), highlight the urgent need for sensitive, deployable tests to diagnose these devastating human diseases. Here we develop CRISPR-Cas13a-based (SHERLOCK) diagnostics targeting Ebola virus (EBOV) and Lassa virus (LASV), with both fluorescent and lateral flow readouts. We demonstrate on laboratory and clinical samples the sensitivity of these assays and the capacity of the SHERLOCK platform to handle virus-specific diagnostic challenges. We perform safety testing to demonstrate the efficacy of our HUDSON protocol in heat-inactivating VHF viruses before SHERLOCK testing, eliminating the need for an extraction. We develop a user-friendly protocol and mobile application (HandLens) to report results, facilitating SHERLOCK's use in endemic regions. Finally, we successfully deploy our tests in Sierra Leone and Nigeria in response to recent outbreaks.
Conflict of interest statement
K.G.B., A.E.L., C.A.F., P.C.S., and C.M. are inventors on patent PCT/US2019/054561 held by the Broad Institute and related to this work. The patent covers all primers, crRNAs, and SHERLOCK technology. P.C.S. is a co-founder of, shareholder in, and advisor to Sherlock Biosciences, Inc., as well as a Board member of and shareholder in Danaher Corporation.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

