DNA methylation mediates differentiation in thermal responses of Pacific oyster (Crassostrea gigas) derived from different tidal levels
- PMID: 32807851
- PMCID: PMC7852555
- DOI: 10.1038/s41437-020-0351-7
DNA methylation mediates differentiation in thermal responses of Pacific oyster (Crassostrea gigas) derived from different tidal levels
Abstract
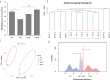
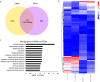
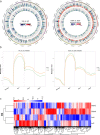
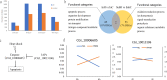
Epigenetic mechanisms such as DNA methylation have the potential to affect organism acclimatization and adaptation to environmental changes by influencing their phenotypic plasticity; however, little is known about the role of methylation in the adaptive phenotypic divergence of marine invertebrates. Therefore, in this study, a typical intertidal species, the Pacific oyster (Crassostrea gigas), was selected to investigate the epigenetic mechanism of phenotypic plasticity in marine invertebrates. Intertidal and subtidal oysters subjected to one-generation common garden experiments and exhibited phenotypic divergence were used. The methylation landscape of both groups of oysters was investigated under temperate and high temperature. The two tidal oysters exhibited divergent methylation patterns, regardless of the temperature, which was mainly original environment-induced. Intertidal samples exhibited significant hypomethylation and more plasticity of methylation in response to heat shock, while subtidal samples showed hypermethylation and less plasticity. Combined with RNA-seq data, a positive relationship between methylation and expression in gene bodies was detected on a genome-wide scale. In addition, approximately 11% and 7% of differentially expressed genes showed significant methylation variation under high temperatures in intertidal and subtidal samples, respectively. Genes related to apoptosis and organism development may be regulated by methylation in response to high temperature in intertidal oysters, whereas oxidation-reduction and ion homeostasis-related genes were involved in subtidal oysters. The results also suggest that DNA methylation mediates phenotypic divergence in oysters adapting to different environments. This study provides new insight into the epigenetic mechanisms underlying phenotypic plasticity in adaptation to rapid climate change in marine organisms.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Andrews S. Babraham bioinformatics. Cambridge, United Kingdom: Babraham Institute; 2010.
-
- Aran D, Toperoff G, Rosenberg M, Hellman A. Replication timing-related and gene body-specific methylation of active human genes. Hum Mol Genet. 2011;20(4):670–680. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

