FAM46C/TENT5C functions as a tumor suppressor through inhibition of Plk4 activity
- PMID: 32807875
- PMCID: PMC7431843
- DOI: 10.1038/s42003-020-01161-3
FAM46C/TENT5C functions as a tumor suppressor through inhibition of Plk4 activity
Abstract
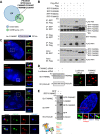
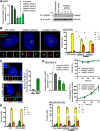
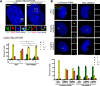
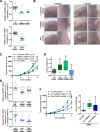
Polo like kinase 4 (Plk4) is a tightly regulated serine threonine kinase that governs centriole duplication. Increased Plk4 expression, which is a feature of many common human cancers, causes centriole overduplication, mitotic irregularities, and chromosomal instability. Plk4 can also promote cancer invasion and metastasis through regulation of the actin cytoskeleton. Herein we demonstrate physical interaction of Plk4 with FAM46C/TENT5C, a conserved protein of unknown function until recently. FAM46C localizes to centrioles, inhibits Plk4 kinase activity, and suppresses Plk4-induced centriole duplication. Interference with Plk4 function by FAM46C was independent of the latter's nucleotidyl transferase activity. In addition, FAM46C restrained cancer cell invasion and suppressed MDA MB-435 cancer growth in a xenograft model, opposing the effect of Plk4. We demonstrate loss of FAM46C in patient-derived colorectal cancer tumor tissue that becomes more profound with advanced clinical stage. These results implicate FAM46C as a tumor suppressor that acts by inhibiting Plk4 activity.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Bettencourt-Dias M, et al. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. CB. 2005;15:2199–2207. - PubMed
-
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 2005;7:1140–1146. - PubMed
-
- Rosario CO, et al. A novel role for Plk4 in regulating cell spreading and motility. Oncogene. 2015;34:3441–3451. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

