ReDU: a framework to find and reanalyze public mass spectrometry data
- PMID: 32807955
- PMCID: PMC7968862
- DOI: 10.1038/s41592-020-0916-7
ReDU: a framework to find and reanalyze public mass spectrometry data
Abstract
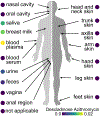
We present ReDU ( https://redu.ucsd.edu/ ), a system for metadata capture of public mass spectrometry-based metabolomics data, with validated controlled vocabularies. Systematic capture of knowledge enables the reanalysis of public data and/or co-analysis of one's own data. ReDU enables multiple types of analyses, including finding chemicals and associated metadata, comparing the shared and different chemicals between groups of samples, and metadata-filtered, repository-scale molecular networking.
Figures