Structure and mechanism of B-family DNA polymerase ζ specialized for translesion DNA synthesis
- PMID: 32807989
- PMCID: PMC7554088
- DOI: 10.1038/s41594-020-0476-7
Structure and mechanism of B-family DNA polymerase ζ specialized for translesion DNA synthesis
Abstract
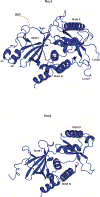
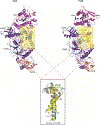
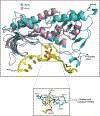
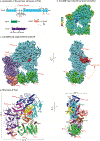
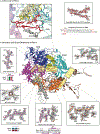
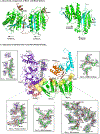
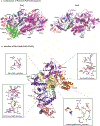
DNA polymerase ζ (Polζ) belongs to the same B-family as high-fidelity replicative polymerases, yet is specialized for the extension reaction in translesion DNA synthesis (TLS). Despite its importance in TLS, the structure of Polζ is unknown. We present cryo-EM structures of the Saccharomyces cerevisiae Polζ holoenzyme in the act of DNA synthesis (3.1 Å) and without DNA (4.1 Å). Polζ displays a pentameric ring-like architecture, with catalytic Rev3, accessory Pol31' Pol32 and two Rev7 subunits forming an uninterrupted daisy chain of protein-protein interactions. We also uncover the features that impose high fidelity during the nucleotide-incorporation step and those that accommodate mismatches and lesions during the extension reaction. Collectively, we decrypt the molecular underpinnings of Polζ's role in TLS and provide a framework for new cancer therapeutics.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures





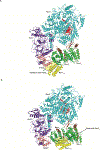


References
-
- Prakash S, Johnson RE & Prakash L Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem 74, 317–353 (2005). - PubMed
-
- Jain R, Aggarwal AK & Rechkoblit O Eukaryotic DNA polymerases. Curr Opin Struct Biol 53, 77–87 (2018). - PubMed
-
- Johnson RE, Washington MT, Haracska L, Prakash S & Prakash L Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature 406, 1015–1019 (2000). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

