Concentration-dependent splicing is enabled by Rbfox motifs of intermediate affinity
- PMID: 32807990
- PMCID: PMC7554199
- DOI: 10.1038/s41594-020-0475-8
Concentration-dependent splicing is enabled by Rbfox motifs of intermediate affinity
Abstract
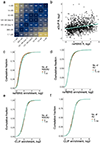
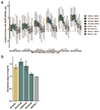
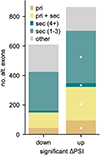
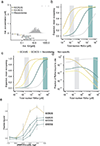
The Rbfox family of splicing factors regulate alternative splicing during animal development and in disease, impacting thousands of exons in the maturing brain, heart and muscle. Rbfox proteins have long been known to bind to the RNA sequence GCAUG with high affinity and specificity, but just half of Rbfox binding sites contain a GCAUG motif in vivo. We incubated recombinant RBFOX2 with over 60,000 mouse and human transcriptomic sequences to reveal substantial binding to several moderate-affinity, non-GCAYG sites at a physiologically relevant range of RBFOX2 concentrations. We find that these 'secondary motifs' bind Rbfox robustly in cells and that several together can exert regulation comparable to GCAUG in a trichromatic splicing reporter assay. Furthermore, secondary motifs regulate RNA splicing in neuronal development and in neuronal subtypes where cellular Rbfox concentrations are highest, enabling a second wave of splicing changes as Rbfox levels increase.
Conflict of interest statement
Competing interests statement
The authors declare no competing interests.
Figures


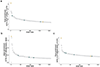









References
-
- Hodgkin J, Zellan JD & Albertson DG Identification of a candidate primary sex determination locus, fox-1, on the X chromosome of Caenorhabditis elegans. Development (1994). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

