Making it or breaking it: DNA methylation and genome integrity
- PMID: 32808652
- PMCID: PMC7606623
- DOI: 10.1042/EBC20200009
Making it or breaking it: DNA methylation and genome integrity
Abstract
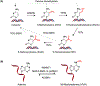
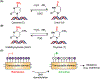
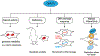
Cells encounter a multitude of external and internal stress-causing agents that can ultimately lead to DNA damage, mutations and disease. A cascade of signaling events counters these challenges to DNA, which is termed as the DNA damage response (DDR). The DDR preserves genome integrity by engaging appropriate repair pathways, while also coordinating cell cycle and/or apoptotic responses. Although many of the protein components in the DDR are identified, how chemical modifications to DNA impact the DDR is poorly understood. This review focuses on our current understanding of DNA methylation in maintaining genome integrity in mammalian cells. DNA methylation is a reversible epigenetic mark, which has been implicated in DNA damage signaling, repair and replication. Sites of DNA methylation can trigger mutations, which are drivers of human diseases including cancer. Indeed, alterations in DNA methylation are associated with increased susceptibility to tumorigenesis but whether this occurs through effects on the DDR, transcriptional responses or both is not entirely clear. Here, we also highlight epigenetic drugs currently in use as therapeutics that target DNA methylation pathways and discuss their effects in the context of the DDR. Finally, we pose unanswered questions regarding the interplay between DNA methylation, transcription and the DDR, positing the potential coordinated efforts of these pathways in genome integrity. While the impact of DNA methylation on gene regulation is widely understood, how this modification contributes to genome instability and mutations, either directly or indirectly, and the potential therapeutic opportunities in targeting DNA methylation pathways in cancer remain active areas of investigation.
Keywords: Cancer; DNA damage; DNA methylation; DNA synthesis and repair; epigenetics; genome integrity.
© 2020 The Author(s). Published by Portland Press Limited on behalf of the Biochemical Society.
Conflict of interest statement
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Figures


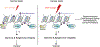
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

