Unlocking Cryptic Metabolites with Mass Spectrometry-Guided Transposon Mutant Selection
- PMID: 32808751
- PMCID: PMC7572862
- DOI: 10.1021/acschembio.0c00558
Unlocking Cryptic Metabolites with Mass Spectrometry-Guided Transposon Mutant Selection
Abstract
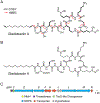
The products of most secondary metabolite biosynthetic gene clusters (BGCs) have yet to be discovered, in part due to low expression levels in laboratory cultures. Reporter-guided mutant selection (RGMS) has recently been developed for this purpose: a mutant library is generated and screened, using genetic reporters to a chosen BGC, to select transcriptionally active mutants that then enable the characterization of the "cryptic" metabolite. The requirement for genetic reporters limits the approach to a single pathway within genetically tractable microorganisms. Herein, we utilize untargeted metabolomics in conjunction with transposon mutagenesis to provide a global read-out of secondary metabolism across large numbers of mutants. We employ self-organizing map analytics and imaging mass spectrometry to identify and characterize seven cryptic metabolites from mutant libraries of two different Burkholderia species. Applications of the methodologies reported can expand our understanding of the products and regulation of cryptic BGCs across phylogenetically diverse bacteria.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
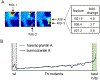
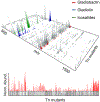
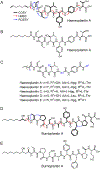
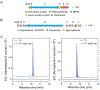
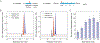
References
-
- Newman DJ, and Cragg GM (2020) Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod 83, 770–803. - PubMed
-
- Nicolaou KC, Sorensen EJ, and Winssinger N (1998) The art and science of organic and natural products synthesis. J. Chem. Educ 75, 1226–1258.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

