The structure-based reaction mechanism of urease, a nickel dependent enzyme: tale of a long debate
- PMID: 32809087
- PMCID: PMC7433671
- DOI: 10.1007/s00775-020-01808-w
The structure-based reaction mechanism of urease, a nickel dependent enzyme: tale of a long debate
Erratum in
-
Correction to: The structure-based reaction mechanism of urease, a nickel dependent enzyme: tale of a long debate.J Biol Inorg Chem. 2021 Feb;26(1):171-173. doi: 10.1007/s00775-021-01855-x. J Biol Inorg Chem. 2021. PMID: 33591411 Free PMC article. No abstract available.
Abstract
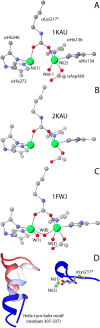
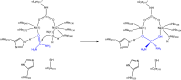
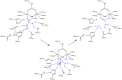
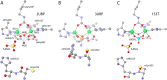
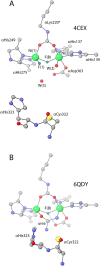
This review is an attempt to retrace the chronicle that starts from the discovery of the role of nickel as the essential metal ion in urease for the enzymatic catalysis of urea, a key step in the biogeochemical cycle of nitrogen on Earth, to the most recent progress in understanding the chemistry of this historical enzyme. Data and facts are presented through the magnifying lenses of the authors, using their best judgment to filter and elaborate on the many facets of the research carried out on this metalloenzyme over the years. The tale is divided in chapters that discuss and describe the results obtained in the subsequent leaps in the knowledge that led from the discovery of a biological role for Ni to the most recent advancements in the comprehension of the relationship between the structure and function of urease. This review is intended not only to focus on the bioinorganic chemistry of this beautiful metal-based catalysis, but also, and maybe primarily, to evoke inspiration and motivation to further explore the realm of bio-based coordination chemistry.
Keywords: Catalytic mechanism; Crystal structure; Helicobacter pylori; Klebsiella aerogenes; Nickel; Sporosarcina pasteurii; Urease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
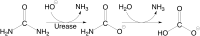
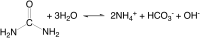
References
-
- Blakeley RL, Hinds JA, Kunze HE, Webb EC, Zerner B. Jack bean urease (EC 3.5.1.5). Demonstration of a carbamoyl-transfer reaction and inhibition by hydroxamic acids. Biochemistry. 1969;8(5):1991–2000. - PubMed
-
- Dixon NE, Riddles PW, Gazzola C, Blakeley RL, Zerner B. Jack bean urease (EC 3.5.1.5). V. On the mechanism of action of urease on urea, formamide, acetamide, N-methylurea, and related compounds. Can J Biochem. 1980;58(12):1335–1344. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources

