Predictions and analyses of RNA nearest neighbor parameters for modified nucleotides
- PMID: 32810273
- PMCID: PMC7498315
- DOI: 10.1093/nar/gkaa654
Predictions and analyses of RNA nearest neighbor parameters for modified nucleotides
Abstract
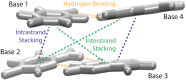
The most popular RNA secondary structure prediction programs utilize free energy (ΔG°37) minimization and rely upon thermodynamic parameters from the nearest neighbor (NN) model. Experimental parameters are derived from a series of optical melting experiments; however, acquiring enough melt data to derive accurate NN parameters with modified base pairs is expensive and time consuming. Given the multitude of known natural modifications and the continuing use and development of unnatural nucleotides, experimentally characterizing all modified NNs is impractical. This dilemma necessitates a computational model that can predict NN thermodynamics where experimental data is scarce or absent. Here, we present a combined molecular dynamics/quantum mechanics protocol that accurately predicts experimental NN ΔG°37 parameters for modified nucleotides with neighboring Watson-Crick base pairs. NN predictions for Watson-Crick and modified base pairs yielded an overall RMSD of 0.32 kcal/mol when compared with experimentally derived parameters. NN predictions involving modified bases without experimental parameters (N6-methyladenosine, 2-aminopurineriboside, and 5-methylcytidine) demonstrated promising agreement with available experimental melt data. This procedure not only yields accurate NN ΔG°37 predictions but also quantifies stacking and hydrogen bonding differences between modified NNs and their canonical counterparts, allowing investigators to identify energetic differences and providing insight into sources of (de)stabilization from nucleotide modifications.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Blind tests of RNA nearest-neighbor energy prediction.Proc Natl Acad Sci U S A. 2016 Jul 26;113(30):8430-5. doi: 10.1073/pnas.1523335113. Epub 2016 Jul 8. Proc Natl Acad Sci U S A. 2016. PMID: 27402765 Free PMC article.
-
Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs.Biochemistry. 1998 Oct 20;37(42):14719-35. doi: 10.1021/bi9809425. Biochemistry. 1998. PMID: 9778347
-
Thermodynamic contribution and nearest-neighbor parameters of pseudouridine-adenosine base pairs in oligoribonucleotides.RNA. 2013 Nov;19(11):1474-82. doi: 10.1261/rna.039610.113. Epub 2013 Sep 23. RNA. 2013. PMID: 24062573 Free PMC article.
-
Thermodynamic parameters for an expanded nearest-neighbor model for the formation of RNA duplexes with single nucleotide bulges.Biochemistry. 2002 Aug 20;41(33):10406-17. doi: 10.1021/bi025781q. Biochemistry. 2002. PMID: 12173927
-
RNA structure and dynamics: a base pairing perspective.Prog Biophys Mol Biol. 2013 Nov;113(2):264-83. doi: 10.1016/j.pbiomolbio.2013.07.003. Epub 2013 Jul 23. Prog Biophys Mol Biol. 2013. PMID: 23891726 Review.
Cited by
-
Secondary structure prediction for RNA sequences including N6-methyladenosine.Nat Commun. 2022 Mar 11;13(1):1271. doi: 10.1038/s41467-022-28817-4. Nat Commun. 2022. PMID: 35277476 Free PMC article.
-
mRNA vaccine sequence and structure design and optimization: Advances and challenges.J Biol Chem. 2025 Jan;301(1):108015. doi: 10.1016/j.jbc.2024.108015. Epub 2024 Nov 26. J Biol Chem. 2025. PMID: 39608721 Free PMC article. Review.
-
Adenine Methylation Enhances the Conformational Flexibility of an RNA Hairpin Tetraloop.J Phys Chem B. 2024 Apr 4;128(13):3157-3166. doi: 10.1021/acs.jpcb.4c00522. Epub 2024 Mar 27. J Phys Chem B. 2024. PMID: 38535997 Free PMC article.
-
Epigenetics of maternal-fetal interface immune microenvironment and placental related pregnancy complications.Front Immunol. 2025 Apr 3;16:1549839. doi: 10.3389/fimmu.2025.1549839. eCollection 2025. Front Immunol. 2025. PMID: 40248704 Free PMC article. Review.
-
A Test and Refinement of Folding Free Energy Nearest Neighbor Parameters for RNA Including N6-Methyladenosine.J Mol Biol. 2022 Sep 30;434(18):167632. doi: 10.1016/j.jmb.2022.167632. Epub 2022 May 16. J Mol Biol. 2022. PMID: 35588868 Free PMC article.
References
-
- Miao Z., Westhof E.. RNA structure: advances and assessment of 3D structure prediction. Annu. Rev. Biophys. 2017; 46:483–503. - PubMed
-
- Preethi S.P., Sharma P., Mitra A.. Higher order structures involving post transcriptionally modified nucleobases in RNA. RSC Adv. 2017; 7:35694–35703.
-
- Li S., Mason C.E.. The pivotal regulatory landscape of RNA modifications. Annu. Rev. Genomics Hum. Genet. 2014; 15:127–150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

