Hepatitis E virus: host tropism and zoonotic infection
- PMID: 32810801
- PMCID: PMC7854786
- DOI: 10.1016/j.mib.2020.07.004
Hepatitis E virus: host tropism and zoonotic infection
Abstract
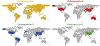
Hepatitis E virus (HEV), the causative agent of hepatitis E, is an understudied but important pathogen. HEV typically causes self-limiting acute viral hepatitis, however chronic infection with neurological and other extrahepatic manifestations has increasingly become a significant clinical problem. The discovery of swine HEV from pigs and demonstration of its zoonotic potential led to the genetic identification of very diverse HEV strains from more than a dozen other animal species. HEV strains from pig, rabbit, deer, camel, and rat have been shown to cross species barriers and infect humans. Zoonotic HEV infections through consumption of raw or undercooked animal meat or direct contact with infected animals have been reported. The discovery of a large number of animal HEV variants does provide an opportunity to develop useful animal models for HEV. In this mini-review, we discuss recent advances in HEV host range, and cross-species and zoonotic transmission.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Conflict of interest
The authors declare no conflict of interest.
Figures

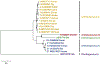

References
-
- Perez-Gracia MT, Suay-Garcia B, Mateos-Lindemann ML: Hepatitis E and pregnancy: current state. Rev Med Virol 2017, 27:e1929. - PubMed
-
- Kamar N, Selves J, Mansuy JM, Ouezzani L, Peron JM, Guitard J, Cointault O, Esposito L, Abravanel F, Danjoux M, et al.: Hepatitis E virus and chronic hepatitis in organ- transplant recipients. N Engl J Med 2008, 358:811–817. - PubMed
-
- Zhang J, Zhang XF, Huang SJ, Wu T, Hu YM, Wang ZZ, Wang H, Jiang HM, Wang YJ, Yan Q, et al.: Long-term efficacy of a hepatitis E vaccine. N Engl J Med 2015, 372:914–922. - PubMed
-
- Kamar N, Izopet J, Tripon S, Bismuth M, Hillaire S, Dumortier J, Radenne S, Coilly A, Garrigue V, D’Alteroche L, et al.: Ribavirin for chronic hepatitis E virus infection in transplant recipients. N Engl J Med 2014, 370:1111–1120. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

