MICAL2 is essential for myogenic lineage commitment
- PMID: 32811811
- PMCID: PMC7434877
- DOI: 10.1038/s41419-020-02886-z
MICAL2 is essential for myogenic lineage commitment
Abstract
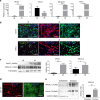
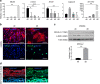
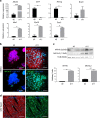
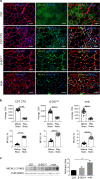
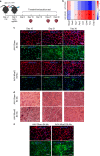
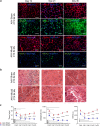
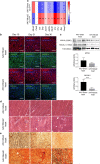
Contractile myofiber units are mainly composed of thick myosin and thin actin (F-actin) filaments. F-Actin interacts with Microtubule Associated Monooxygenase, Calponin And LIM Domain Containing 2 (MICAL2). Indeed, MICAL2 modifies actin subunits and promotes actin filament turnover by severing them and preventing repolymerization. In this study, we found that MICAL2 increases during myogenic differentiation of adult and pluripotent stem cells (PSCs) towards skeletal, smooth and cardiac muscle cells and localizes in the nucleus of acute and chronic regenerating muscle fibers. In vivo delivery of Cas9-Mical2 guide RNA complexes results in muscle actin defects and demonstrates that MICAL2 is essential for skeletal muscle homeostasis and functionality. Conversely, MICAL2 upregulation shows a positive impact on skeletal and cardiac muscle commitments. Taken together these data demonstrate that modulations of MICAL2 have an impact on muscle filament dynamics and its fine-tuned balance is essential for the regeneration of muscle tissues.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
References
-
- Zammit PS. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin Cell Dev. Biol. 2017;72:19–32. - PubMed
-
- Camps J, et al. Interstitial cell remodeling promotes aberrant adipogenesis in dystrophic muscles. Cell Rep. 2020;31:107597. - PubMed
-
- Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 2004;84:209–238. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

