Prevention of dsRNA-induced interferon signaling by AGO1x is linked to breast cancer cell proliferation
- PMID: 32812257
- PMCID: PMC7507497
- DOI: 10.15252/embj.2019103922
Prevention of dsRNA-induced interferon signaling by AGO1x is linked to breast cancer cell proliferation
Abstract
Translational readthrough, i.e., elongation of polypeptide chains beyond the stop codon, was initially reported for viral RNA, but later found also on eukaryotic transcripts, resulting in proteome diversification and protein-level modulation. Here, we report that AGO1x, an evolutionarily conserved translational readthrough isoform of Argonaute 1, is generated in highly proliferative breast cancer cells, where it curbs accumulation of double-stranded RNAs (dsRNAs) and consequent induction of interferon responses and apoptosis. In contrast to other mammalian Argonaute protein family members with primarily cytoplasmic functions, AGO1x exhibits nuclear localization in the vicinity of nucleoli. We identify AGO1x interaction with the polyribonucleotide nucleotidyltransferase 1 (PNPT1) and show that the depletion of this protein further augments dsRNA accumulation. Our study thus uncovers a novel function of an Argonaute protein in buffering the endogenous dsRNA-induced interferon responses, different than the canonical function of AGO proteins in the miRNA effector pathway. As AGO1x expression is tightly linked to breast cancer cell proliferation, our study thus suggests a new direction for limiting tumor growth.
Keywords: Argonaute 1; breast cancer; endogenous dsRNA; interferon response; translation readthrough.
© 2020 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

- A
Top: schema of analyzed TR regions (purple), located downstream of the annotated open reading frame (gray), between the annotated stop codon (red triangle) and the next in‐frame stop codon (orange triangle); Bottom: histogram of average PhastCons conservation scores (x‐axis) of putative TR regions of all RefSeq‐annotated transcripts. The scores of the four human Argonaute protein family members are highlighted.
- B
Multiple sequence alignment of the AGO1 putative TR region across vertebrates.
- C
Multiple sequence alignment of the corresponding predicted amino acid sequence. The unique peptide targeted by the polyclonal antibody is indicated by the red line. The green and blue lines indicate peptide sequences obtained after tryptic digestion, in which cleavage is exclusively at arginine (R) and lysine (K) (further described below in panels E and F). Red asterisks indicate stop codons.
- D
Western blot showing AGO1x expression in three cell lines. For comparison, a parallel blot was probed with an antibody directed against canonical AGO1. Tubulin served as loading control.
- E, F
Annotated MS/MS spectrum of peptides specific for the endogenous AGO1x, “QNAVTSLDR”, depicted in green (E) and “LSKPQELCHPNPEEAR”, depicted in blue (F). The Mascot ion score (text color corresponds to peptides marked in Fig 1C for reference) as well as the annotated fragments (blue = y‐ions; red = b‐ions) together with the corresponding amino acids is indicated.

- A
Western blot analysis of multiple cell lines demonstrates that in addition to the canonical AGO1 protein band, a characteristic second band of higher MW is revealed by the AGO1 antibody.
- B
Higher MW band observed in (A) corresponds to AGO1x protein. Representative Western blot shows that the intensity of the higher MW band is sensitive to ectopic overexpression (using the pIRES‐Neo vector) of FLAG‐tagged AGO1x but not of FLAG‐tagged AGO1. Expression of the corresponding isoform is confirmed with a blot for FLAG expression. The overexpression constructs are indicated with labels above the blots. Protein ladders show that the higher MW band corresponds to approximately 100 kDa.
- C, D
Western blot with AGO1x antibody demonstrates its specificity for the AGO1x isoform stably expressed from pCDH‐FLAG-tagged plasmids. Middle and lower panels depict AGO1x levels in individual samples, at low and high exposure, respectively, of the same blot. The higher exposure was used to better assess the expression level in untransfected (control) samples.
- E, F
Representative images of MDA‐MB-231 stained with AGO1 (green) and AGO1x (red) antibodies under conditions of endogenous expression or knockdown with an siRNA pool targeting the transcript that encodes both isoforms (E). An AGO1x overexpression system, where FLAG‐AGO1x was stably integrated into MDA‐MB-231 cells, was also tested (F). DAPI was used to mark the nucleus (blue).
- G
Western blot analysis of AGO1x protein level in cells treated with either siAGO1 or siControl confirms the imaging results from panel E.

Representative immunofluorescence images showing the subcellular distribution of AGO1x (red) relative to nuclear and cytoplasmic markers. DAPI was used to mark the nucleus (blue). The co‐stained subcellular marker is indicated in each panel in green. SC 35, Lsm4, α‐tubulin, ERP72, p54 (NRB), and nucleolin serve as markers for nuclear speckles, cytosol and nucleus, cytosol, endoplasmic reticulum, paraspeckle, and nucleolus, respectively.
Mean (± SD) pixel intensities of AGO1x staining in nucleolus and nucleoplasm, computed from z‐stack images of MDA‐MB-231 cells (n = 20). The P‐value was determined using a paired two‐tailed t‐test.
Representative AGO1x and AGO1 blot from MDA-MB‐231 cell fractions. GAPDH and hnRNP C1/C2 served as markers of purity of the individual fractions.
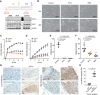
Top: scheme of the sites targeted by sgRNA in the generation of AGO1x mutant cell lines. Bottom: Western blot analysis of lysates from mutant MDA-MB‐231 cells showing the depletion of AGO1x and the comparable total AGO1 levels across the cell lines. Three independently collected sets of cells were lysed and analyzed in parallel. After calculating the levels of AGO1x and AGO1 relative to GAPDH, values corresponding to individual lanes were normalized to the average over the replicates of the control line. These numbers are indicated below the blots. The P‐values of the t‐tests comparing the expression AGO1x in mutant (W1A and W6A) and control (CTRL) lines were W1A‐CTRL: 0.0008 and W6A‐CTRL: 0.0002; and for AGO1, W1A‐CTRL: 0.1227 and W6A‐CTRL: 0.1173.
Phase‐contrast images of control and mutant cell lines at 24 and 48 h after plating equal numbers (10,000) of cells in individual wells of a six‐well plate.
Impedance‐based mean (± SD) cell indices at the indicated time points after seeding equal numbers of CTRL (n = 6), W1A (n = 5), and W6A (n = 5) cells. From 6 h on, there is a statistically significant difference between control and the two mutant cell lines (P < 0.001, two‐tailed t‐test).
Impedance‐based mean (± SD) cell indices as a function of time for control (n = 3), W1A (n = 4), and W6A (n = 3) cells grown in electronically monitored Boyden chambers. From 12 h on, there is a statistical significant difference between control and the two mutants (P < 0.005, two‐tailed t‐test).
Mean (± SD) number of colonies obtained in a soft agar colony formation assay of control, W1A, and W6A mutant cell lines (all n = 4, P‐value determined using an unpaired two‐tailed t‐test).
Mean (± SD) number of spheres obtained from a sphere formation assay of control, W1A, and W6A mutant cell lines (all n = 8, P‐values computed as in E).
AGO1x staining of tissue sections from breast cancers, classified as low proliferating or high proliferating based on Ki‐67 staining. The upper and lower panel represents two different tissue sections.
Mean (± SD) percentage of AGO1x‐positive cells in breast tumors with low (n = 7) and high (n = 9) proliferation index, assessed by the expression level of Ki‐67. P‐value was determined using an unpaired two‐tailed t‐test.

Representative images of colony formation assays performed with the W1A, W6A, and control MDA-MB‐231 cell lines. Images were captured with an inverted microscope (ZEISS Axio Vert. A1 equipped with an AxioCam MRc camera).
Representative images of sphere formation assays performed with W1A, W6A, and control MDA-MB‐231 cell lines. Images were captured with an inverted phase‐contrast microscope (Leica) and were inverted for improved contrast.
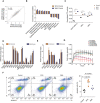
Scatter plot of mean log2 mRNA fold changes in the mutant cell lines versus control (n = 3). Shown are also the Pearson correlation coefficient and respective P‐value. The dashed line indicates equal change in the two mutant lines.
Normalized enrichment score (ES) from gene set enrichment analysis (Subramanian et al, 2005) comparing gene expression changes in the mutant cell lines relative to the control cell line. For all depicted pathways, P < 0.05. P‐values were calculated by comparing the empirical ES of a gene set to a null distribution of ESs derived from permuting the gene set and then adjusted for multiple hypothesis testing.
Mean (± SD) activity of STAT1/IRF1/IRF2/IRF3/IRF9 transcription factor motifs estimated by ISMARA (Balwierz et al, 2014) (n = 3). Shown are the P‐values (***P < 0.001) determined by Dunn's multiple comparison test post hoc and the non‐parametric Kruskal–Wallis test (P < 0.0001).
Mean (± SEM) log2 mRNA expression fold changes (inferred from RNA‐seq) of genes involved in the interferon‐alpha response and apoptosis in the two mutant cell lines relative to control (n = 3). Multiple testing‐corrected P‐values for Wald tests comparing fold changes with respect to control are depicted above each bar (**P < 0.01, ***P < 0.001).
Mean (± SD) expression fold changes of the corresponding proteins (if detected in the proteomics data) in the two mutant cell lines (W1A, n = 2; W6A, n = 1) compared to control (n = 2). Multiple testing‐corrected P‐values for unpaired two‐tailed t‐tests (proteomics) comparing fold changes with respect to control are depicted above each bar (*P < 0.05, **P < 0.01, ***P < 0.001).
Representative result of the apoptosis assay using annexin V and propidium iodide (PI) staining in the control and the two mutant cell lines (left). The percentage of cells in each quadrant is depicted for each cell line. Quantification of the mean (± SD) percentage of apoptotic cells (Q1+Q2, Annexin V+) across the different cell lines (n = 4) (right). Shown is the P‐value determined by unpaired two‐tailed t‐test.
Impedance‐based mean (± SD) cell index values at the indicated time points of growth after seeding equal numbers of control, W1A, and W6A cells, upon treatment with DMSO or ruxolitinib. For the DMSO treatment: control (n = 4), W1A (n = 3), and W6A (n = 3). For the ruxolitinib treatment: control (n = 6), W1A (n = 3), and W6A (n = 4). From 6 h after DMSO treatment, there is a statistically significant difference between control and the two mutants (P < 0.05, two‐tailed t‐test), whereas no statistically significant difference is found after ruxolitinib treatment.
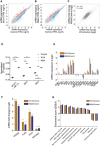
- A, B
Scatter plots of mean log10 mRNA expression levels (in transcript counts per million, TPM) in W1A (A) and W6A mutant cell lines (B) compared to the control line (n = 3). mRNAs that are significantly upregulated or downregulated (|fold change| > 2 and FDR < 0.01) in the mutant cell lines are shown in red and blue, respectively. The dashed line indicates equal mRNA levels for control and mutant cell lines.
- C
Scatter plot of mean log2 mRNA fold changes from three biological replicates in the two mutant cell lines compared to control. Shown is also the Pearson correlation coefficient and respective P‐value. Dashed line indicates equal change in the two mutant lines.
- D
Mean (± SD) activity of STAT1/IRF1/IRF2/IRF3/IRF9 transcription factor motifs estimated by ISMARA (Balwierz et al, 2014) from the RNA‐seq data (n = 3). Shown are the P‐values (***P < 0.001) determined by Dunn's multiple comparison test post hoc and the non‐parametric Kruskal–Wallis test (P < 0.0001).
- E, F
Mean (± SEM) log2 mRNA expression fold changes of genes involved in the interferon‐alpha response and apoptosis (E) and in dsRNA sensing (F) in the two mutant cell lines relative to control (n = 3). Multiple testing‐corrected P‐values for fold changes with respect to control are depicted above each bar (*P < 0.05, **P < 0.01, ***P < 0.001).
- G
Normalized enrichment score (ES) from gene set enrichment analysis comparing gene expression in the two mutant cell lines with that in the control cell line. For all the pathways depicted, P < 0.05. P‐values were calculated by comparing the empirical ES of a gene set relative to a null distribution of ESs derived from permuting the gene set and then adjusted for multiple hypothesis testing.

GFP fluorescent microscopy images of control and mutant cell lines infected at MOI of 0.1 or 1 with SINV‐GFP virus for 48 h. The left panel corresponds to the GFP signal from infected cells and the right panel to a merge of GFP signal and bright field. Pictures were taken with a 5× magnification. MOI: multiplicity of infection; hpi: hours post‐infection.
Representative Western blot of p‐PKR, p‐eIF2alpha (Ser‐52), and GFP expression in SINV‐GFP-infected cells in the same condition as in (A). Tubulin was used as loading control.
Mean (± SEM) of SINV‐GFP viral titers in control and mutant lines infected at an MOI of 0.1 or 1 for 48 h (n = 3) from plaque assay quantification. **P < 0.001. P‐value was obtained using an ordinary one‐way ANOVA test comparing mutant cell lines to control.
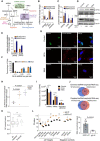
- A
Schematic representation of the dsRNA sensing mechanism, downstream signaling events, and expected phenotypes.
- B
Mean (± SEM) log2 mRNA fold changes of dsRNAs sensors (from panel A) in the two mutant cell lines compared to control (n = 3) inferred from RNA‐seq. Multiple testing‐corrected P‐values for Wald tests comparing fold changes with respect to control are depicted above each bar (***P < 0.001).
- C
Mean (± SD) fold changes of corresponding proteins (see B), if detected in the proteomics data from the two mutant (W1A, n = 2; W6A, n = 1) lines relative to control (n = 2). Multiple testing‐corrected P‐values for unpaired two‐tailed t‐tests (proteomics) comparing fold changes with respect to control are depicted above each bar (*P < 0.05, **P < 0.01, ***P < 0.001).
- D
Western blot detection of phosphorylated eIF2α (Ser51) and total eIF2α levels in cell lysates of control and mutant MDA-MB‐231 cells. GAPDH and tubulin serve as loading control for phosphorylated eIF2α and total eIF2α blots, respectively. Numbers indicated below the blots represent the band intensities of target proteins normalized to the loading controls, with the value in the control line taken as baseline of 1.
- E
Mean (± SD) fold changes in phosphorylated peptide (phosphorylation sites are indicated in brackets) levels derived from a proteomic analysis of phosphopeptides in the two mutant lines (W1A, n = 2; W6A, n = 2) compared to control (n = 1). Multiple testing‐corrected P‐values for Wald tests (transcriptomics) or unpaired two‐tailed t‐tests (proteomics) comparing fold changes with respect to control are depicted above each bar (**P < 0.01).
- F
Quantification of the relative abundance (mean ± SEM) of distinct dsRNA foci in each cell type. 3D images were projected on a single plane, non‐overlapping foci were counted from 100 cells of each type, and the numbers in three abundance bins (< 1, 1–5, > 5) are shown.
- G
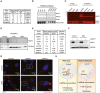
Representative immunofluorescence image of the control and mutant MDA-MB‐231 cells stained with AGO1x (green) and J2 antibody (red) and DAPI (blue) to mark the nucleus. Loss of AGO1x signal in mutants also confirms the efficacy of CRISPR‐induced mutations.
- H, I
Concentration of recovered RNA (mean ± SEM) measured by NanoDrop™ (H) and relative abundance of various RNA species (mean ± SD) in J2 antibody immunoprecipitates from three biological replicates of control and mutant cell lines (I). Multiple testing‐corrected P‐values from unpaired two‐tailed t‐tests are depicted above each comparison (*P < 0.05, ***P < 0.001).
- J
Venn diagrams showing the intersection between RNA transcripts that were consistently enriched (top) or depleted (bottom) in the dsRNA‐seq relative to total RNA‐seq in the two mutant cell lines compared to control (n = 3).
- K
Boxplots showing the proportion of G/C nucleotides in all genes and in genes depleted/enriched in dsRNA‐seq compared to total RNA‐seq (n = 3). Shown are the P‐values in the non‐parametric Mann–Whitney U test (***P < 0.001). Boxes extend from the 25th to 75th percentiles (inter‐quartile range (IQR)), horizontal lines represent the median, whiskers indicate the lowest and highest data within 1.5×IQR from the lower and upper quartiles, respectively.
- L
Left: quantification of transcript abundance (mean ± SEM) by qRT–PCR in AGO1x (black, n = 6 for putative targets, n = 3 for negative controls) or IgG (orange, n = 2) IP from control cells. n.d., not detected. Multiple testing‐corrected P‐values for an unpaired two‐tailed t‐test are depicted above each comparison (*P < 0.05, **P < 0.01, ***P < 0.001). Right: concentration of RNA (mean ± SD) obtained with each type of antibody, with P‐value from an unpaired two‐tailed Student's t‐test.
- A
Summary of mass spectrometric analysis of AGO1x (n = 5) or IgG (n = 2) IP from control cells. P‐values were calculated using Fisher's exact test and corrected for multiple testing using the Benjamini–Hochberg method.
- B
Western blot analysis to validate the interaction of AGO1x with PNPT1 and DHX9 in control MDA-MB‐231 cells. AGO1x pulldowns were compared to IgG pulldowns to verify the relative levels of PNPT1 and DHX9 from four independent biological replicates. The immunoglobulin heavy chain band served as a loading control.
- C, D
Validation of AGO1x‐PNPT1 interaction by reciprocal pulldown. Immunoprecipitation of PNPT1 from control and mutant MDA-MB‐231 cells and Western blotting to validate the specificity of PNPT1 antibody in comparison with IgG control (C). Presence of AGO1x and fibrillarin in PNPT1 immunoprecipitates. Three independent biological replicates were used, and the eluted proteins were analyzed by Western blotting (D).
- E
In vitro interaction assay of isolated nuclear extracts from control MDA-MB‐231 cells with FLAG‐tagged AGO1 or FLAG‐tagged AGO1x immunoprecipitated from cells stably expressing the constructs. Table indicates the peptide counts from a second round of immunoprecipitation with anti‐FLAG antibody and subsequent analysis by LC‐MS/MS. P‐values are from Fisher's exact test, corrected for multiple testing using the Benjamini–Hochberg method.
- F, G
Western blot analysis to verify the depletion of PNPT1 upon siRNA treatment (F) and a representative immunofluorescence image of siPNPT1/siControl‐treated control and W6A mutant MDA-MB‐231 cells stained with J2 antibody (red). DAPI was used to mark the nucleus (blue) (G). The right‐most panels show a magnification of the cells enclosed by the white boxes in the middle panels.
- H
Model of AGO1x function. A complex of AGO1x with PNPT1 interacts with nuclear RNAs to prevent the accumulation of structured dsRNAs, supporting proliferation of breast cancer cells. Depletion of AGO1x leads to deleterious accumulation of these RNAs, which in turn leads to activation of interferon (IFN) response and apoptosis.

- A–F
Representative micrographs of AGO1x expression in normal breast tissue (A), lung parenchyma (B), colon mucosa (C), kidney tissue (D), stomach mucosa (E), and prostate tissue (F). Two representative non‐tumoral tissue samples are shown for each organ. Insets with high magnification images highlight different degrees of positivity (red arrows) in the six different non‐tumoral tissues. Scale bars, 100 μm.
Similar articles
-
Let-7a-regulated translational readthrough of mammalian AGO1 generates a microRNA pathway inhibitor.EMBO J. 2019 Aug 15;38(16):e100727. doi: 10.15252/embj.2018100727. Epub 2019 Jul 22. EMBO J. 2019. PMID: 31330067 Free PMC article.
-
Function of HNRNPC in breast cancer cells by controlling the dsRNA-induced interferon response.EMBO J. 2018 Dec 3;37(23):e99017. doi: 10.15252/embj.201899017. Epub 2018 Aug 29. EMBO J. 2018. PMID: 30158112 Free PMC article.
-
Argonaute 2 drives miR-145-5p-dependent gene expression program in breast cancer cells.Cell Death Dis. 2019 Jan 8;10(1):17. doi: 10.1038/s41419-018-1267-5. Cell Death Dis. 2019. PMID: 30622242 Free PMC article.
-
Seeking the truth behind the myth: Argonaute tales from "nuclearland".Mol Cell. 2022 Feb 3;82(3):503-513. doi: 10.1016/j.molcel.2021.11.005. Epub 2021 Dec 1. Mol Cell. 2022. PMID: 34856122 Review.
-
Host defense, viruses and apoptosis.Cell Death Differ. 2001 Feb;8(2):113-26. doi: 10.1038/sj.cdd.4400823. Cell Death Differ. 2001. PMID: 11313713 Review.
Cited by
-
Activity and Function in Human Cells of the Evolutionary Conserved Exonuclease Polynucleotide Phosphorylase.Int J Mol Sci. 2022 Jan 31;23(3):1652. doi: 10.3390/ijms23031652. Int J Mol Sci. 2022. PMID: 35163574 Free PMC article. Review.
-
Signaling Through Nucleic Acid Sensors and Their Roles in Inflammatory Diseases.Front Immunol. 2021 Jan 28;11:625833. doi: 10.3389/fimmu.2020.625833. eCollection 2020. Front Immunol. 2021. PMID: 33633744 Free PMC article. Review.
-
DNA Hypomethylation Underlies Epigenetic Swapping between AGO1 and AGO1-V2 Isoforms in Tumors.Epigenomes. 2024 Jun 22;8(3):24. doi: 10.3390/epigenomes8030024. Epigenomes. 2024. PMID: 39051182 Free PMC article.
-
Argonaute Proteins: From Structure to Function in Development and Pathological Cell Fate Determination.Front Cell Dev Biol. 2020 Jan 22;7:360. doi: 10.3389/fcell.2019.00360. eCollection 2019. Front Cell Dev Biol. 2020. PMID: 32039195 Free PMC article. Review.
-
Response to Suresh et al.EMBO J. 2025 Jul;44(14):3920-3922. doi: 10.1038/s44318-025-00479-0. Epub 2025 Jun 11. EMBO J. 2025. PMID: 40500329 Free PMC article.
References
-
- Ahrné E, Glatter T, Viganò C, von Schubert C, Nigg EA, Schmidt A (2016) Evaluation and improvement of quantification accuracy in isobaric mass tag‐based protein quantification experiments. J Proteome Res 15: 2537–2547 - PubMed
-
- Ameyar‐Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau J‐C et al (2012) Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol 19: 998–1004 - PubMed
-
- Atienza JM, Zhu J, Wang X, Xu X, Abassi Y (2005) Dynamic monitoring of cell adhesion and spreading on microelectronic sensor arrays. J Biomol Screen 10: 795–805 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

