Paneth Cell-Derived Lysozyme Defines the Composition of Mucolytic Microbiota and the Inflammatory Tone of the Intestine
- PMID: 32814028
- PMCID: PMC7461615
- DOI: 10.1016/j.immuni.2020.07.010
Paneth Cell-Derived Lysozyme Defines the Composition of Mucolytic Microbiota and the Inflammatory Tone of the Intestine
Abstract
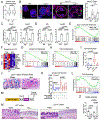
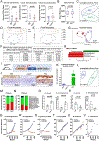
Paneth cells are the primary source of C-type lysozyme, a β-1,4-N-acetylmuramoylhydrolase that enzymatically processes bacterial cell walls. Paneth cells are normally present in human cecum and ascending colon, but are rarely found in descending colon and rectum; Paneth cell metaplasia in this region and aberrant lysozyme production are hallmarks of inflammatory bowel disease (IBD) pathology. Here, we examined the impact of aberrant lysozyme production in colonic inflammation. Targeted disruption of Paneth cell lysozyme (Lyz1) protected mice from experimental colitis. Lyz1-deficiency diminished intestinal immune responses to bacterial molecular patterns and resulted in the expansion of lysozyme-sensitive mucolytic bacteria, including Ruminococcus gnavus, a Crohn's disease-associated pathobiont. Ectopic lysozyme production in colonic epithelium suppressed lysozyme-sensitive bacteria and exacerbated colitis. Transfer of R. gnavus into Lyz1-/- hosts elicited a type 2 immune response, causing epithelial reprograming and enhanced anti-colitogenic capacity. In contrast, in lysozyme-intact hosts, processed R. gnavus drove pro-inflammatory responses. Thus, Paneth cell lysozyme balances intestinal anti- and pro-inflammatory responses, with implications for IBD.
Keywords: IL-13; Lyz1; Paneth cell; Ruminococcus gnavus; colitis; inflammation; lysozyme; mucolytic bacteria; type 2 immunity.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures





Comment in
-
Lysozyme: A Double-Edged Sword in the Intestine.Trends Immunol. 2020 Dec;41(12):1054-1056. doi: 10.1016/j.it.2020.10.010. Epub 2020 Nov 3. Trends Immunol. 2020. PMID: 33158739
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- F31 DK121428/DK/NIDDK NIH HHS/United States
- R01 DK093668/DK/NIDDK NIH HHS/United States
- F32 CA235829/CA/NCI NIH HHS/United States
- R01 DK109711/DK/NIDDK NIH HHS/United States
- R21 CA178599/CA/NCI NIH HHS/United States
- R01 AI134040/AI/NIAID NIH HHS/United States
- R01 DK102934/DK/NIDDK NIH HHS/United States
- R01 DK119198/DK/NIDDK NIH HHS/United States
- R01 AT010243/AT/NCCIH NIH HHS/United States
- R01 HL123340/HL/NHLBI NIH HHS/United States
- R01 CA190558/CA/NCI NIH HHS/United States
- K99 CA245123/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

