Transcription and DNA Methylation Patterns of Blood-Derived CD8+ T Cells Are Associated With Age and Inflammatory Bowel Disease But Do Not Predict Prognosis
- PMID: 32814113
- PMCID: PMC7428744
- DOI: 10.1053/j.gastro.2020.08.017
Transcription and DNA Methylation Patterns of Blood-Derived CD8+ T Cells Are Associated With Age and Inflammatory Bowel Disease But Do Not Predict Prognosis
Abstract
Background & aims: Gene expression patterns of CD8+ T cells have been reported to correlate with clinical outcomes of adults with inflammatory bowel diseases (IBD). We aimed to validate these findings in independent patient cohorts.
Methods: We obtained peripheral blood samples from 112 children with a new diagnosis of IBD (71 with Crohn's disease and 41 with ulcerative colitis) and 19 children without IBD (controls) and recorded medical information on disease activity and outcomes. CD8+ T cells were isolated from blood samples by magnetic bead sorting at the point of diagnosis and during the course of disease. Genome-wide transcription (n = 192) and DNA methylation (n = 66) profiles were generated using Affymetrix and Illumina arrays, respectively. Publicly available transcriptomes and DNA methylomes of CD8+ T cells from 3 adult patient cohorts with and without IBD were included in data analyses.
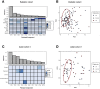
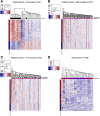
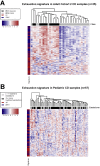
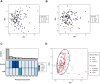
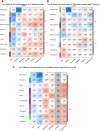
Results: Previously reported CD8+ T-cell prognostic expression and exhaustion signatures were only found in the original adult IBD patient cohort. These signatures could not be detected in either a pediatric or a second adult IBD cohort. In contrast, an association between CD8+ T-cell gene expression with age and sex was detected across all 3 cohorts. CD8+ gene transcription was clearly associated with IBD in the 2 cohorts that included non-IBD controls. Lastly, DNA methylation profiles of CD8+ T cells from children with Crohn's disease correlated with age but not with disease outcome.
Conclusions: We were unable to validate previously reported findings of an association between CD8+ T-cell gene transcription and disease outcome in IBD. Our findings reveal the challenges of developing prognostic biomarkers for patients with IBD and the importance of their validation in large, independent cohorts before clinical application.
Keywords: Biomarker; Epigenetic; Prognosis; Validation.
Copyright © 2021 AGA Institute. Published by Elsevier Inc. All rights reserved.
Figures




Comment in
-
Deciphering the role of CD8+ T cells in IBD: from single-cell analysis to biomarkers.Nat Rev Gastroenterol Hepatol. 2020 Oct;17(10):595. doi: 10.1038/s41575-020-00362-9. Nat Rev Gastroenterol Hepatol. 2020. PMID: 32884120 No abstract available.
-
Validation of Predictive Models in Inflammatory Bowel Diseases.Gastroenterology. 2021 Jan;160(1):21-22. doi: 10.1053/j.gastro.2020.11.005. Epub 2020 Nov 12. Gastroenterology. 2021. PMID: 33188805 No abstract available.
-
Analytical Mistakes Confound Attempted Validation: A Response to "Transcription and DNA Methylation Patterns of Blood-Derived CD8+ T Cells Are Associated With Age and Inflammatory Bowel Disease But Do Not Predict Prognosis".Gastroenterology. 2021 May;160(6):2210-2211. doi: 10.1053/j.gastro.2021.01.021. Epub 2021 Jan 13. Gastroenterology. 2021. PMID: 33453231 No abstract available.
-
Reply.Gastroenterology. 2021 May;160(6):2211-2212. doi: 10.1053/j.gastro.2021.01.212. Epub 2021 Jan 25. Gastroenterology. 2021. PMID: 33508288 No abstract available.
References
-
- Herzog D., Fournier N., Buehr P. Age at disease onset of inflammatory bowel disease is associated with later extraintestinal manifestations and complications. Eur J Gastroenterol Hepatol. 2018;30:598–607. - PubMed
-
- Gravano D.M., Hoyer K.K. Promotion and prevention of autoimmune disease by CD8+ T cells. J Autoimmun. 2013;45:68–79. - PubMed
Supplementary References
-
- Davis S., Meltzer P.S. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

