On-target versus off-target effects of drugs inhibiting the replication of SARS-CoV-2
- PMID: 32814759
- PMCID: PMC7434849
- DOI: 10.1038/s41419-020-02842-x
On-target versus off-target effects of drugs inhibiting the replication of SARS-CoV-2
Abstract
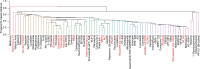
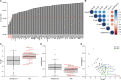
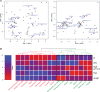
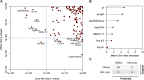
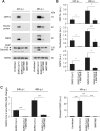
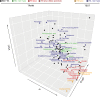
The current epidemic of coronavirus disease-19 (COVID-19) caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) calls for the development of inhibitors of viral replication. Here, we performed a bioinformatic analysis of published and purported SARS-CoV-2 antivirals including imatinib mesylate that we found to suppress SARS-CoV-2 replication on Vero E6 cells and that, according to the published literature on other coronaviruses is likely to act on-target, as a tyrosine kinase inhibitor. We identified a cluster of SARS-CoV-2 antivirals with characteristics of lysosomotropic agents, meaning that they are lipophilic weak bases capable of penetrating into cells. These agents include cepharentine, chloroquine, chlorpromazine, clemastine, cloperastine, emetine, hydroxychloroquine, haloperidol, ML240, PB28, ponatinib, siramesine, and zotatifin (eFT226) all of which are likely to inhibit SARS-CoV-2 replication by non-specific (off-target) effects, meaning that they probably do not act on their 'official' pharmacological targets, but rather interfere with viral replication through non-specific effects on acidophilic organelles including autophagosomes, endosomes, and lysosomes. Imatinib mesylate did not fall into this cluster. In conclusion, we propose a tentative classification of SARS-CoV-2 antivirals into specific (on-target) versus non-specific (off-target) agents based on their physicochemical characteristics.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
References
-
- Guha R. Chemical informatics functionality in R. 2007. 2007;18:16.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

