NLRP7 plays a functional role in regulating BMP4 signaling during differentiation of patient-derived trophoblasts
- PMID: 32814763
- PMCID: PMC7438493
- DOI: 10.1038/s41419-020-02884-1
NLRP7 plays a functional role in regulating BMP4 signaling during differentiation of patient-derived trophoblasts
Abstract
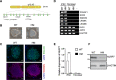
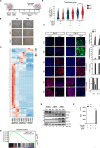
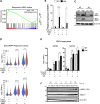
Complete hydatidiform mole (HM) is a gestational trophoblastic disease resulting in hyperproliferation of trophoblast cells and absence of embryo development. Mutations in the maternal-effect gene NLRP7 are the major cause of familial recurrent complete HM. Here, we established an in vitro model of HM using patient-specific induced pluripotent stem cells (iPSCs) derived trophoblasts harboring NLRP7 mutations. Using whole transcriptome profiling during trophoblast differentiation, we showed that impaired NLRP7 expression results in precocious downregulation of pluripotency factors, activation of trophoblast lineage markers, and promotes maturation of differentiated extraembryonic cell types such as syncytiotrophoblasts. Interestingly, we found that these phenotypes are dependent on BMP4 signaling and BMP pathway inhibition corrected the excessive trophoblast differentiation of patient-derived iPSCs. Our human iPSC model of a genetic placental disease recapitulates aspects of trophoblast biology, highlights the broad utility of iPSC-derived trophoblasts for modeling human placental diseases and identifies NLRP7 as an essential modulator of key developmental cell fate regulators.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Li R, Albertini DF. The road to maturation: somatic cell interaction and self-organization of the mammalian oocyte. Nat. Rev. Mol. Cell Biol. 2013;14:141–152. - PubMed
-
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. - PubMed
-
- Lu X, Gao Z, Qin D, Li L. A maternal functional module in the mammalian oocyte-to-embryo transition. Trends Mol. Med. 2017;23:1014–1023. - PubMed
-
- Hui P, Buza N, Murphy KM, Ronnett BM. Hydatidiform moles: genetic basis and precision diagnosis. Annu. Rev. Pathol. Mech. Dis. 2017;12:449–485. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

