Histone methyltransferase DOT1L coordinates AR and MYC stability in prostate cancer
- PMID: 32814769
- PMCID: PMC7438336
- DOI: 10.1038/s41467-020-18013-7
Histone methyltransferase DOT1L coordinates AR and MYC stability in prostate cancer
Abstract
The histone methyltransferase DOT1L methylates lysine 79 (K79) on histone H3 and is involved in Mixed Lineage Leukemia (MLL) fusion leukemogenesis; however, its role in prostate cancer (PCa) is undefined. Here we show that DOT1L is overexpressed in PCa and is associated with poor outcome. Genetic and chemical inhibition of DOT1L selectively impaired the viability of androgen receptor (AR)-positive PCa cells and organoids, including castration-resistant and enzalutamide-resistant cells. The sensitivity of AR-positive cells is due to a distal K79 methylation-marked enhancer in the MYC gene bound by AR and DOT1L not present in AR-negative cells. DOT1L inhibition leads to reduced MYC expression and upregulation of MYC-regulated E3 ubiquitin ligases HECTD4 and MYCBP2, which promote AR and MYC degradation. This leads to further repression of MYC in a negative feed forward manner. Thus DOT1L selectively regulates the tumorigenicity of AR-positive prostate cancer cells and is a promising therapeutic target for PCa.
Conflict of interest statement
The authors declare no competing interests.
Figures



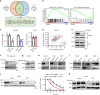


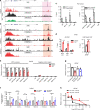

Comment in
-
DOT1L in prostate cancer.Nat Rev Urol. 2020 Oct;17(10):544. doi: 10.1038/s41585-020-0374-0. Nat Rev Urol. 2020. PMID: 32895552 No abstract available.
-
Uro-Science.J Urol. 2021 May;205(5):1517-1519. doi: 10.1097/JU.0000000000001658. Epub 2021 Feb 24. J Urol. 2021. PMID: 33625915 No abstract available.
References
-
- Mohammad HP, Barbash O, Creasy CL. Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat. Med. 2019;25:403–418. - PubMed
-
- Feng Q, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 2002;12:1052–1058. - PubMed
-
- Huyen Y, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. - PubMed
-
- Okada Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. - PubMed
-
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum. Mol. Genet. 2007;16:92–106. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

