Nearest-neighbor amino acids of specificity-determining residues influence the activity of engineered Cre-type recombinases
- PMID: 32814809
- PMCID: PMC7438526
- DOI: 10.1038/s41598-020-70867-5
Nearest-neighbor amino acids of specificity-determining residues influence the activity of engineered Cre-type recombinases
Abstract
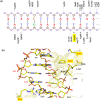
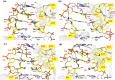
The tyrosine-type site-specific DNA recombinase Cre recombines its target site, loxP, with high activity and specificity without cross-recombining the target sites of highly related recombinases. Understanding how Cre achieves this precision is key to be able to rationally engineer site-specific recombinases (SSRs) for genome editing applications. Previous work has revealed key residues for target site selectivity in the Cre/loxP and the related Dre/rox recombinase systems. However, enzymes in which these residues were changed to the respective counterpart only showed weak activity on the foreign target site. Here, we use molecular modeling and dynamics simulation techniques to comprehensively explore the mechanisms by which these residues determine target recognition in the context of their flanking regions in the protein-DNA interface, and we establish a structure-based rationale for the design of improved recombination activities. Our theoretical models reveal that nearest-neighbors to the specificity-determining residues are important players for enhancing SSR activity on the foreign target site. Based on the established rationale, we design new Cre variants with improved rox recombination activities, which we validate experimentally. Our work provides new insights into the target recognition mechanisms of Cre-like recombinases and represents an important step towards the rational design of SSRs for applied genome engineering.
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Structure and mechanism in site-specific recombination.Curr Opin Struct Biol. 1999 Feb;9(1):14-20. doi: 10.1016/s0959-440x(99)80003-7. Curr Opin Struct Biol. 1999. PMID: 10047575 Review.
-
Discovery of Nigri/nox and Panto/pox site-specific recombinase systems facilitates advanced genome engineering.Sci Rep. 2016 Jul 22;6:30130. doi: 10.1038/srep30130. Sci Rep. 2016. PMID: 27444945 Free PMC article.
-
Single molecule TPM analysis of the catalytic pentad mutants of Cre and Flp site-specific recombinases: contributions of the pentad residues to the pre-chemical steps of recombination.Nucleic Acids Res. 2015 Mar 31;43(6):3237-55. doi: 10.1093/nar/gkv114. Epub 2015 Mar 12. Nucleic Acids Res. 2015. PMID: 25765648 Free PMC article.
-
Chimeras of the Flp and Cre recombinases: tests of the mode of cleavage by Flp and Cre.J Mol Biol. 2000 Sep 8;302(1):27-48. doi: 10.1006/jmbi.2000.3967. J Mol Biol. 2000. PMID: 10964559
-
Cre Recombinase and Other Tyrosine Recombinases.Chem Rev. 2016 Oct 26;116(20):12785-12820. doi: 10.1021/acs.chemrev.6b00077. Epub 2016 May 10. Chem Rev. 2016. PMID: 27163859 Review.
Cited by
-
Activation of recombinases at specific DNA loci by zinc-finger domain insertions.Nat Biotechnol. 2024 Dec;42(12):1844-1854. doi: 10.1038/s41587-023-02121-y. Epub 2024 Jan 31. Nat Biotechnol. 2024. PMID: 38297187 Free PMC article.
-
A Novel Cre/lox71-Based System for Inducible Expression of Recombinant Proteins and Genome Editing.Cells. 2022 Jul 7;11(14):2141. doi: 10.3390/cells11142141. Cells. 2022. PMID: 35883584 Free PMC article.
-
Precise excision of HTLV-1 provirus with a designer-recombinase.Mol Ther. 2023 Jul 5;31(7):2266-2285. doi: 10.1016/j.ymthe.2023.03.014. Epub 2023 Mar 17. Mol Ther. 2023. PMID: 36934299 Free PMC article.
-
Pairing of single mutations yields obligate Cre-type site-specific recombinases.Nucleic Acids Res. 2022 Jan 25;50(2):1174-1186. doi: 10.1093/nar/gkab1240. Nucleic Acids Res. 2022. PMID: 34951450 Free PMC article.
-
Prediction of designer-recombinases for DNA editing with generative deep learning.Nat Commun. 2022 Dec 27;13(1):7966. doi: 10.1038/s41467-022-35614-6. Nat Commun. 2022. PMID: 36575171 Free PMC article.
References
-
- Grindley NDF, Whiteson KL, Rice PA. Mechanisms of Site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. - PubMed
-
- Jayaram M, Ma CH, Kachroo AH, Rowley PA, Guga P, Fan HF. An overview of tyrosine site-specific recombination: from an flp perspective. Microbiol Spectr. 2015;3:43–71.. - PubMed
-
- Kilby NJ, Snaith MR, Murray JAH. Site-specific recombinases: tools for genome engineering. Trends Genet. 1993;9:413–421. - PubMed
-
- Meinke G, Bohm A, Hauber J, Pisabarro MT, Buchholz F. Cre recombinase and other tyrosine recombinases. Chem. Rev. 2016;116:12785–12820. - PubMed
-
- Van Duyne GD. A structural view of Cre- loxP site-specific recombination. Annu. Rev. Biophys. Biomol. Struct. 2001;30:87–104. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

