Genetic and functional insights into the fractal structure of the heart
- PMID: 32814899
- PMCID: PMC7116759
- DOI: 10.1038/s41586-020-2635-8
Genetic and functional insights into the fractal structure of the heart
Abstract
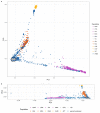
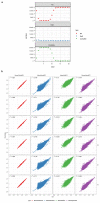
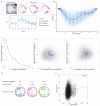
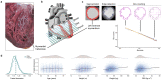
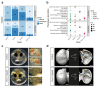
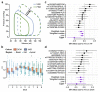
The inner surfaces of the human heart are covered by a complex network of muscular strands that is thought to be a remnant of embryonic development1,2. The function of these trabeculae in adults and their genetic architecture are unknown. Here we performed a genome-wide association study to investigate image-derived phenotypes of trabeculae using the fractal analysis of trabecular morphology in 18,096 participants of the UK Biobank. We identified 16 significant loci that contain genes associated with haemodynamic phenotypes and regulation of cytoskeletal arborization3,4. Using biomechanical simulations and observational data from human participants, we demonstrate that trabecular morphology is an important determinant of cardiac performance. Through genetic association studies with cardiac disease phenotypes and Mendelian randomization, we find a causal relationship between trabecular morphology and risk of cardiovascular disease. These findings suggest a previously unknown role for myocardial trabeculae in the function of the adult heart, identify conserved pathways that regulate structural complexity and reveal the influence of the myocardial trabeculae on susceptibility to cardiovascular disease.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Sizarov A, et al. Formation of the building plan of the human heart: morphogenesis, growth, and differentiation. Circulation. 2011;123:1125–35. - PubMed
-
- Kawabata Galbraith K, et al. MTSS1 regulation of actin-nucleating formin DAAM1 in dendritic filopodia determines final dendritic configuration of Purkinje cells. Cell Rep. 2018;24:95–106.e9. - PubMed
Publication types
MeSH terms
Grants and funding
- RE/18/4/34215/BHF_/British Heart Foundation/United Kingdom
- MC_UP_1102/19/MRC_/Medical Research Council/United Kingdom
- MC_PC_17114/MRC_/Medical Research Council/United Kingdom
- R01ES029917-02/ES/NIEHS NIH HHS/United States
- RG/19/6/34387/BHF_/British Heart Foundation/United Kingdom
- MR/M003191/1/MRC_/Medical Research Council/United Kingdom
- P30 CA045508/CA/NCI NIH HHS/United States
- NH/17/1/32725/BHF_/British Heart Foundation/United Kingdom
- MC_U120085815/MRC_/Medical Research Council/United Kingdom
- 107469/WT_/Wellcome Trust/United Kingdom
- MR/N026934/1/MRC_/Medical Research Council/United Kingdom
- G9901399/MRC_/Medical Research Council/United Kingdom
- G9409531/MRC_/Medical Research Council/United Kingdom
- G0900897/MRC_/Medical Research Council/United Kingdom
- G9409634/MRC_/Medical Research Council/United Kingdom
- MC_UP_1102/20/MRC_/Medical Research Council/United Kingdom
- 107469/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- R01 ES029917/ES/NIEHS NIH HHS/United States
- MR/K501013/1/MRC_/Medical Research Council/United Kingdom
- MR/S003754/1/MRC_/Medical Research Council/United Kingdom
- SP/17/11/32885/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources

