Genetic Studies of Hypertrophic Cardiomyopathy in Singaporeans Identify Variants in TNNI3 and TNNT2 That Are Common in Chinese Patients
- PMID: 32815737
- PMCID: PMC7676617
- DOI: 10.1161/CIRCGEN.119.002823
Genetic Studies of Hypertrophic Cardiomyopathy in Singaporeans Identify Variants in TNNI3 and TNNT2 That Are Common in Chinese Patients
Abstract
Background: To assess the genetic architecture of hypertrophic cardiomyopathy (HCM) in patients of predominantly Chinese ancestry.
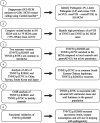
Methods: We sequenced HCM disease genes in Singaporean patients (n=224) and Singaporean controls (n=3634), compared findings with additional populations and White HCM cohorts (n=6179), and performed in vitro functional studies.
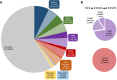
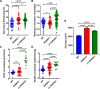
Results: Singaporean HCM patients had significantly fewer confidently interpreted HCM disease variants (pathogenic/likely pathogenic: 18%, P<0.0001) but an excess of variants of uncertain significance (24%, P<0.0001), as compared to Whites (pathogenic/likely pathogenic: 31%, excess of variants of uncertain significance: 7%). Two missense variants in thin filament encoding genes were commonly seen in Singaporean HCM (TNNI3:p.R79C, disease allele frequency [AF]=0.018; TNNT2:p.R286H, disease AF=0.022) and are enriched in Singaporean HCM when compared with Asian controls (TNNI3:p.R79C, Singaporean controls AF=0.0055, P=0.0057, genome aggregation database-East Asian AF=0.0062, P=0.0086; TNNT2:p.R286H, Singaporean controls AF=0.0017, P<0.0001, genome aggregation database-East Asian AF=0.0009, P<0.0001). Both these variants have conflicting annotations in ClinVar and are of low penetrance (TNNI3:p.R79C, 0.7%; TNNT2:p.R286H, 2.7%) but are predicted to be deleterious by computational tools. In population controls, TNNI3:p.R79C carriers had significantly thicker left ventricular walls compared with noncarriers while its etiological fraction is limited (0.70 [95% CI, 0.35-0.86]) and thus TNNI3:p.R79C is considered variant of uncertain significance. Mutant TNNT2:p.R286H iPSC-CMs (induced pluripotent stem cells derived cardiomyocytes) show hypercontractility, increased metabolic requirements, and cellular hypertrophy and the etiological fraction (0.93 [95% CI, 0.83-0.97]) support the likely pathogenicity of TNNT2:p.R286H.
Conclusions: As compared with Whites, Chinese HCM patients commonly have low penetrance risk alleles in TNNT2 or TNNI3 but exhibit few clinically actionable HCM variants overall. This highlights the need for greater study of HCM genetics in non-White populations.
Keywords: cardiomyopathies; hypertrophy; population; troponin I; troponin T.
Figures

References
-
- Maron BJ. Hypertrophic cardiomyopathy: a systematic review. JAMA. 2002;287:1308–1320. doi: 10.1001/jama.287.10.1308 - PubMed
-
- Elliott PM, Anastasakis A, Borger MA, Borggrefe M, Cecchi F, Charron P, Hagege AA, Lafont A, Limongelli G, Mahrholdt H, et al. ; Authors/Task Force members. 2014 ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy: the task force for the diagnosis and management of hypertrophic cardiomyopathy of the European Society of Cardiology (ESC). Eur Heart J. 2014;35:2733–79. doi: 10.1093/eurheartj/ehu284 - PubMed
-
- Jarcho JA, McKenna W, Pare JA, Solomon SD, Holcombe RF, Dickie S, Levi T, Donis-Keller H, Seidman JG, Seidman CE. Mapping a gene for familial hypertrophic cardiomyopathy to chromosome 14q1. N Engl J Med. 1989;321:1372–1378. doi: 10.1056/NEJM198911163212005 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

