The sps Genes Encode an Original Legionaminic Acid Pathway Required for Crust Assembly in Bacillus subtilis
- PMID: 32817102
- PMCID: PMC7439481
- DOI: 10.1128/mBio.01153-20
The sps Genes Encode an Original Legionaminic Acid Pathway Required for Crust Assembly in Bacillus subtilis
Abstract
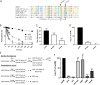
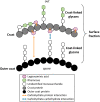
The crust is the outermost spore layer of most Bacillus strains devoid of an exosporium. This outermost layer, composed of both proteins and carbohydrates, plays a major role in the adhesion and spreading of spores into the environment. Recent studies have identified several crust proteins and have provided insights about their organization at the spore surface. However, although carbohydrates are known to participate in adhesion, little is known about their composition, structure, and localization. In this study, we showed that the spore surface of Bacillus subtilis is covered with legionaminic acid (Leg), a nine-carbon backbone nonulosonic acid known to decorate the flagellin of the human pathogens Helicobacter pylori and Campylobacter jejuni We demonstrated that the spsC, spsD, spsE, spsG, and spsM genes of Bacillus subtilis are required for Leg biosynthesis during sporulation, while the spsF gene is required for Leg transfer from the mother cell to the surface of the forespore. We also characterized the activity of SpsM and highlighted an original Leg biosynthesis pathway in B. subtilis Finally, we demonstrated that Leg is required for the assembly of the crust around the spores, and we showed that in the absence of Leg, spores were more adherent to stainless steel probably because of their reduced hydrophilicity and charge.IMPORTANCEBacillus species are a major economic and food safety concern of the food industry because of their food spoilage-causing capability and persistence. Their persistence is mainly due to their ability to form highly resistant spores adhering to the surfaces of industrial equipment. Spores of the Bacillus subtilis group are surrounded by the crust, a superficial layer which plays a key role in their adhesion properties. However, knowledge of the composition and structure of this layer remains incomplete. Here, for the first time, we identified a nonulosonic acid (Leg) at the surfaces of bacterial spores (B. subtilis). We uncovered a novel Leg biosynthesis pathway, and we demonstrated that Leg is required for proper crust assembly. This work contributes to the description of the structure and composition of Bacillus spores which has been under way for decades, and it provides keys to understanding the importance of carbohydrates in Bacillus adhesion and persistence in the food industry.
Keywords: Bacillus subtilis; bacterial adhesion; crust; legionaminic acid; nonulosonic acid; spores.
Copyright © 2020 Dubois et al.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

