Alternative N-terminal regions of Drosophila myosin heavy chain II regulate communication of the purine binding loop with the essential light chain
- PMID: 32817166
- PMCID: PMC7573276
- DOI: 10.1074/jbc.RA120.014684
Alternative N-terminal regions of Drosophila myosin heavy chain II regulate communication of the purine binding loop with the essential light chain
Abstract
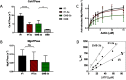
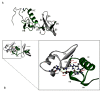
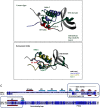
We investigated the biochemical and biophysical properties of one of the four alternative exon-encoded regions within the Drosophila myosin catalytic domain. This region is encoded by alternative exons 3a and 3b and includes part of the N-terminal β-barrel. Chimeric myosin constructs (IFI-3a and EMB-3b) were generated by exchanging the exon 3-encoded areas between native slow embryonic body wall (EMB) and fast indirect flight muscle myosin isoforms (IFI). We found that this exchange alters the kinetic properties of the myosin S1 head. The ADP release rate (k-D ) in the absence of actin is completely reversed for each chimera compared with the native isoforms. Steady-state data also suggest a reciprocal shift, with basal and actin-activated ATPase activity of IFI-3a showing reduced values compared with wild-type (WT) IFI, whereas for EMB-3b these values are increased compared with wild-type (WT) EMB. In the presence of actin, ADP affinity (KAD ) is unchanged for IFI-3a, compared with IFI, but ADP affinity for EMB-3b is increased, compared with EMB, and shifted toward IFI values. ATP-induced dissociation of acto-S1 (K1k+2 ) is reduced for both exon 3 chimeras. Homology modeling, combined with a recently reported crystal structure for Drosophila EMB, indicates that the exon 3-encoded region in the myosin head is part of the communication pathway between the nucleotide binding pocket (purine binding loop) and the essential light chain, emphasizing an important role for this variable N-terminal domain in regulating actomyosin crossbridge kinetics, in particular with respect to the force-sensing properties of myosin isoforms.
Keywords: actin; fluorescence; force-sensing; homology modeling; kinetics; muscle; myosin; protein structure-function; sequence alignment.
© 2020 Bloemink et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures






References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

