The XPO6 Exportin Mediates Herpes Simplex Virus 1 gM Nuclear Release Late in Infection
- PMID: 32817212
- PMCID: PMC7565615
- DOI: 10.1128/JVI.00753-20
The XPO6 Exportin Mediates Herpes Simplex Virus 1 gM Nuclear Release Late in Infection
Abstract
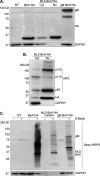
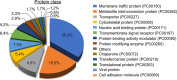
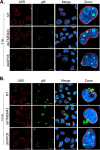
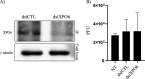
The glycoprotein M of herpes simplex virus 1 (HSV-1) is dynamically relocated from nuclear membranes to the trans-Golgi network (TGN) during infection, but molecular partners that promote this relocalization are unknown. Furthermore, while the presence of the virus is essential for this phenomenon, it is not clear if this is facilitated by viral or host proteins. Past attempts to characterize glycoprotein M (gM) interacting partners identified the viral protein gN by coimmunoprecipitation and the host protein E-Syt1 through a proteomics approach. Interestingly, both proteins modulate the activity of gM on the viral fusion machinery. However, neither protein is targeted to the nuclear membrane and consequently unlikely explains the dynamic regulation of gM nuclear localization. We thus reasoned that gM may transiently interact with other molecules. To resolve this issue, we opted for a proximity-dependent biotin identification (BioID) proteomics approach by tagging gM with a BirA* biotinylation enzyme and purifying BirA substrates on a streptavidin column followed by mass spectrometry analysis. The data identified gM and 170 other proteins that specifically and reproducibly were labeled by tagged gM at 4 or 12 h postinfection. Surprisingly, 35% of these cellular proteins are implicated in protein transport. Upon testing select candidate proteins, we discovered that XPO6, an exportin, is required for gM to be released from the nucleus toward the TGN. This is the first indication of a host or viral protein that modulates the presence of HSV-1 gM on nuclear membranes.IMPORTANCE The mechanisms that enable integral proteins to be targeted to the inner nuclear membrane are poorly understood. Herpes simplex virus 1 (HSV-1) glycoprotein M (gM) is an interesting candidate, as it is dynamically relocalized from nuclear envelopes to the trans-Golgi network (TGN) in a virus- and time-dependent fashion. However, it was, until now, unclear how gM was directed to the nucleus or evaded that compartment later on. Through a proteomic study relying on a proximity-ligation assay, we identified several novel gM interacting partners, many of which are involved in vesicular transport. Analysis of select proteins revealed that XPO6 is required for gM to leave the nuclear membranes late in the infection. This was unexpected, as XPO6 is an exportin specifically associated with actin/profilin nuclear export. This raises some very interesting questions about the interaction of HSV-1 with the exportin machinery and the cargo specificity of XPO6.
Keywords: BioID; INM; RANBP20; Ran; Ran-binding protein 20; UL10; XPO6; exportin; herpes; herpes simplex; herpesvirus; host-pathogen interactions; inner nuclear membrane; intracellular transport; nuclear egress.
Copyright © 2020 American Society for Microbiology.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

