DDX5 promotes oncogene C3 and FABP1 expressions and drives intestinal inflammation and tumorigenesis
- PMID: 32817263
- PMCID: PMC7441524
- DOI: 10.26508/lsa.202000772
DDX5 promotes oncogene C3 and FABP1 expressions and drives intestinal inflammation and tumorigenesis
Abstract
Tumorigenesis in different segments of the intestinal tract involves tissue-specific oncogenic drivers. In the colon, complement component 3 (C3) activation is a major contributor to inflammation and malignancies. By contrast, tumorigenesis in the small intestine involves fatty acid-binding protein 1 (FABP1). However, little is known of the upstream mechanisms driving their expressions in different segments of the intestinal tract. Here, we report that the RNA-binding protein DDX5 binds to the mRNA transcripts of C3 and Fabp1 to augment their expressions posttranscriptionally. Knocking out DDX5 in epithelial cells protected mice from intestinal tumorigenesis and dextran sodium sulfate (DSS)-induced colitis. Identification of DDX5 as a common upstream regulator of tissue-specific oncogenic molecules provides an excellent therapeutic target for intestinal diseases.
© 2020 Abbasi et al.
Conflict of interest statement
GW Yeo is co-founder, member of the Board of Directors, on the Science Advisory Board, equity holder, and paid consultant for Locanabio and Eclipse BioInnovations. GW Yeo is a visiting professor at the National University of Singapore. GW Yeo’s interests have been reviewed and approved by the University of California San Diego in accordance with its conflict of interest policies. All other authors declare that they have no conflict of interest.
Figures






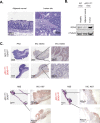





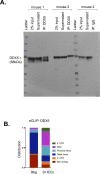

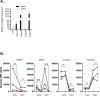
References
-
- Leon F, Contractor N, Fuss I, Marth T, Lahey E, Iwaki S, la Sala A, Hoffmann V, Strober W, Kelsall BL (2006) Antibodies to complement receptor 3 treat established inflammation in murine models of colitis and a novel model of psoriasiform dermatitis. J Immunol 177: 6974–6982. 10.4049/jimmunol.177.10.6974 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UH3 TR002968/TR/NCATS NIH HHS/United States
- R01 CA238042/CA/NCI NIH HHS/United States
- R01 CA100768/CA/NCI NIH HHS/United States
- U41 HG009889/HG/NHGRI NIH HHS/United States
- R01 DK107585/DK/NIDDK NIH HHS/United States
- R01 HG004659/HG/NHGRI NIH HHS/United States
- R01 GM138385/GM/NIGMS NIH HHS/United States
- UG3 TR002968/TR/NCATS NIH HHS/United States
- T32 CA067754/CA/NCI NIH HHS/United States
- UG3 TR003355/TR/NCATS NIH HHS/United States
- R00 CA151673/CA/NCI NIH HHS/United States
- S10 OD026929/OD/NIH HHS/United States
- R01 AI141630/AI/NIAID NIH HHS/United States
- P30 CA023100/CA/NCI NIH HHS/United States
- R01 GM124494/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
