A three-gene signature based on MYC, BCL-2 and NFKBIA improves risk stratification in diffuse large B-cell lymphoma
- PMID: 32817282
- PMCID: PMC8409021
- DOI: 10.3324/haematol.2019.236455
A three-gene signature based on MYC, BCL-2 and NFKBIA improves risk stratification in diffuse large B-cell lymphoma
Abstract
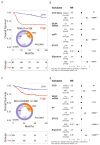
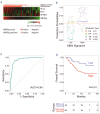
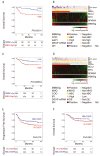
Recent randomized trials focused on gene expression-based determination of the cell of origin in diffuse large B-cell lymphoma could not show significant improvements by adding novel agents to standard chemoimmunotherapy. The aim of this study was the identification of a gene signature able to refine current prognostication algorithms and applicable to clinical practice. Here we used a targeted gene expression profiling panel combining the Lymph2Cx signature for cell of origin classification with additional targets including MYC, BCL-2 and NFKBIA, in 186 patients from 2 randomized trials (discovery cohort) (NCT00355199 and NCT00499018). Data were validated in 3 independent series (2 large public datasets and a real-life cohort). By integrating the cell of origin, MYC/BCL-2 double expressor status and NFKBIA expression, we defined a 3-gene signature combining MYC, BCL-2 and NFKBIA (MBN-signature), which outperformed the MYC/BCL-2 double expressor status in multivariate analysis, and allowed further risk stratification within the germinal center B-cell/unclassified subset. The high-risk (MBN Sig-high) subgroup identified the vast majority of double hit cases and a significant fraction of Activated B-Cell-derived diffuse large B-cell lymphomas. These results were validated in 3 independent series including a cohort from the REMoDL-B trial, where, in an exploratory ad hoc analysis, the addition of bortezomib in the MBN Sig-high subgroup provided a progression free survival advantage compared with standard chemoimmunotherapy. These data indicate that a simple 3-gene signature based on MYC, BCL-2 and NFKBIA could refine the prognostic stratification in diffuse large B-cell lymphoma, and might be the basis for future precision-therapy approaches.
Figures


References
-
- Alizadeh AA, Eisen MB, Davis RE, et al. . Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503-511. - PubMed
-
- Shipp MA, Ross KN, Tamayo P, et al. . Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8(1):68-74. - PubMed
-
- Hans CP, Weisenburger DD, Greiner TC, et al. . Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275-282. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

