A comparison of four serological assays for detecting anti-SARS-CoV-2 antibodies in human serum samples from different populations
- PMID: 32817357
- PMCID: PMC7665313
- DOI: 10.1126/scitranslmed.abc3103
A comparison of four serological assays for detecting anti-SARS-CoV-2 antibodies in human serum samples from different populations
Abstract
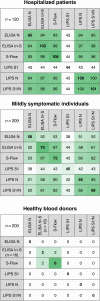
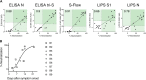
It is of paramount importance to evaluate the prevalence of both asymptomatic and symptomatic cases of SARS-CoV-2 infection and their differing antibody response profiles. Here, we performed a pilot study of four serological assays to assess the amounts of anti-SARS-CoV-2 antibodies in serum samples obtained from 491 healthy individuals before the SARS-CoV-2 pandemic, 51 individuals hospitalized with COVID-19, 209 suspected cases of COVID-19 with mild symptoms, and 200 healthy blood donors. We used two ELISA assays that recognized the full-length nucleoprotein (N) or trimeric spike (S) protein ectodomain of SARS-CoV-2. In addition, we developed the S-Flow assay that recognized the S protein expressed at the cell surface using flow cytometry, and the luciferase immunoprecipitation system (LIPS) assay that recognized diverse SARS-CoV-2 antigens including the S1 domain and the carboxyl-terminal domain of N by immunoprecipitation. We obtained similar results with the four serological assays. Differences in sensitivity were attributed to the technique and the antigen used. High anti-SARS-CoV-2 antibody titers were associated with neutralization activity, which was assessed using infectious SARS-CoV-2 or lentiviral-S pseudotype virus. In hospitalized patients with COVID-19, seroconversion and virus neutralization occurred between 5 and 14 days after symptom onset, confirming previous studies. Seropositivity was detected in 32% of mildly symptomatic individuals within 15 days of symptom onset and in 3% of healthy blood donors. The four antibody assays that we used enabled a broad evaluation of SARS-CoV-2 seroprevalence and antibody profiling in different subpopulations within one region.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures



References
-
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E. C., Zhang Y.-Z., A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). - PMC - PubMed
-
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). - PMC - PubMed
-
- Corman V. M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D. K. W., Bleicker T., Brünink S., Schneider J., Schmidt M. L., Mulders D. G. J. C., Haagmans B. L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M. P. G., Drosten C., Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 25, 2000045 (2020). - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

