Targeting acid ceramidase inhibits YAP/TAZ signaling to reduce fibrosis in mice
- PMID: 32817366
- PMCID: PMC7976849
- DOI: 10.1126/scitranslmed.aay8798
Targeting acid ceramidase inhibits YAP/TAZ signaling to reduce fibrosis in mice
Abstract
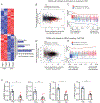
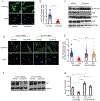
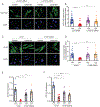
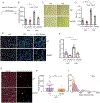
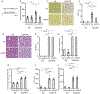
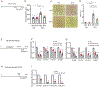
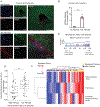
Hepatic stellate cells (HSCs) drive hepatic fibrosis. Therapies that inactivate HSCs have clinical potential as antifibrotic agents. We previously identified acid ceramidase (aCDase) as an antifibrotic target. We showed that tricyclic antidepressants (TCAs) reduce hepatic fibrosis by inhibiting aCDase and increasing the bioactive sphingolipid ceramide. We now demonstrate that targeting aCDase inhibits YAP/TAZ activity by potentiating its phosphorylation-mediated proteasomal degradation via the ubiquitin ligase adaptor protein β-TrCP. In mouse models of fibrosis, pharmacologic inhibition of aCDase or genetic knockout of aCDase in HSCs reduces fibrosis, stromal stiffness, and YAP/TAZ activity. In patients with advanced fibrosis, aCDase expression in HSCs is increased. Consistently, a signature of the genes most down-regulated by ceramide identifies patients with advanced fibrosis who could benefit from aCDase targeting. The findings implicate ceramide as a critical regulator of YAP/TAZ signaling and HSC activation and highlight aCDase as a therapeutic target for the treatment of fibrosis.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures
References
-
- Li Z, Dranoff JA, Chan EP, Uemura M, Sevigny J, Wells RG, Transforming growth factor-beta and substrate stiffness regulate portal fibroblast activation in culture. Hepatology 46, 1246–1256 (2007). - PubMed
-
- Georges PC, Hui JJ, Gombos Z, McCormick ME, Wang AY, Uemura M, Mick R, Janmey PA, Furth EE, Wells RG, Increased stiffness of the rat liver precedes matrix deposition: implications for fibrosis. Am J Physiol Gastrointest Liver Physiol 293, G1147–1154 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

