Tracking of Antibiotic Resistance Transfer and Rapid Plasmid Evolution in a Hospital Setting by Nanopore Sequencing
- PMID: 32817379
- PMCID: PMC7440845
- DOI: 10.1128/mSphere.00525-20
Tracking of Antibiotic Resistance Transfer and Rapid Plasmid Evolution in a Hospital Setting by Nanopore Sequencing
Abstract
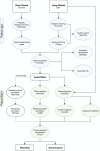
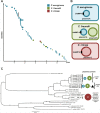
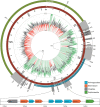
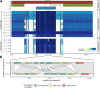
Infections with multidrug-resistant bacteria often leave limited or no treatment options. The transfer of antimicrobial resistance genes (ARG) carrying plasmids between bacterial species by horizontal gene transfer represents an important mode of expansion of ARGs. Here, we demonstrate the application of Nanopore sequencing in a hospital setting for monitoring transfer and rapid evolution of antibiotic resistance plasmids within and across multiple species. In 2009, we experienced an outbreak with extensively multidrug-resistant Pseudomonas aeruginosa harboring the carbapenemase-encoding blaIMP-8 gene. In 2012, the first Citrobacter freundii and Citrobacter cronae strains harboring the same gene were detected. Using Nanopore and Illumina sequencing, we conducted comparative analysis of all blaIMP-8 bacteria isolated in our hospital over a 6-year period (n = 54). We developed the computational platform plasmIDent for Nanopore-based characterization of clinical isolates and monitoring of ARG transfer, comprising de novo assembly of genomes and plasmids, plasmid circularization, ARG annotation, comparative genome analysis of multiple isolates, and visualization of results. Using plasmIDent, we identified a 40-kb plasmid carrying blaIMP-8 in P. aeruginosa and C. freundii, verifying the plasmid transfer. Within C. freundii, the plasmid underwent further evolution and plasmid fusion, resulting in a 164-kb megaplasmid, which was transferred to C. cronae Multiple rearrangements of the multidrug resistance gene cassette were detected in P. aeruginosa, including deletions and translocations of complete ARGs. In summary, plasmid transfer, plasmid fusion, and rearrangement of the ARG cassette mediated the rapid evolution of opportunistic pathogens in our hospital. We demonstrated the feasibility of near-real-time monitoring of plasmid evolution and ARG transfer in clinical settings, enabling successful countermeasures to contain plasmid-mediated outbreaks.IMPORTANCE Infections with multidrug-resistant bacteria represent a major threat to global health. While the spread of multidrug-resistant bacterial clones is frequently studied in the hospital setting, surveillance of the transfer of mobile genetic elements between different bacterial species was difficult until recent advances in sequencing technologies. Nanopore sequencing technology was applied to track antimicrobial gene transfer in a long-term outbreak of multidrug-resistant Pseudomonas aeruginosa, Citrobacter freundii, and Citrobacter cronae in a German hospital over 6 years. We developed a novel computational pipeline, pathoLogic, which enables de novo assembly of genomes and plasmids, antimicrobial resistance gene annotation and visualization, and comparative analysis. Applying this approach, we detected plasmid transfer between different bacterial species as well as plasmid fusion and frequent rearrangements of the antimicrobial resistance gene cassette. This study demonstrated the feasibility of near-real-time tracking of plasmid-based antimicrobial resistance gene transfer in hospitals, enabling countermeasures to contain plasmid-mediated outbreaks.
Keywords: IMP-8; Nanopore; Nanopore sequencing; Pseudomonas aeruginosa; antimicrobial resistance; genome assembly; horizontal gene transfer; long read; pathoLogic; plasmIDent; plasmid-mediated resistance; plasmids; surveillance studies.
Copyright © 2020 Peter et al.
Figures
References
-
- Roca I, Akova M, Baquero F, Carlet J, Cavaleri M, Coenen S, Cohen J, Findlay D, Gyssens I, Heure OE, Kahlmeter G, Kruse H, Laxminarayan R, Liébana E, López-Cerero L, MacGowan A, Martins M, Rodríguez-Baño J, Rolain J-M, Segovia C, Sigauque B, Tacconelli E, Wellington E, Vila J. 2015. The global threat of antimicrobial resistance: science for intervention. New Microbes New Infect 6:22–29. doi: 10.1016/j.nmni.2015.02.007. - DOI - PMC - PubMed
-
- Kengkla K, Kongpakwattana K, Saokaew S, Apisarnthanarak A, Chaiyakunapruk N. 2018. Comparative efficacy and safety of treatment options for MDR and XDR Acinetobacter baumannii infections: a systematic review and network meta-analysis. J Antimicrob Chemother 73:22–32. doi: 10.1093/jac/dkx368. - DOI - PubMed
-
- Tacconelli E, Cataldo MA, Dancer SJ, De Angelis G, Falcone M, Frank U, Kahlmeter G, Pan A, Petrosillo N, Rodriguez-Bano J, Singh N, Venditti M, Yokoe DS, Cookson B, European Society of Clinical M. 2014. ESCMID guidelines for the management of the infection control measures to reduce transmission of multidrug-resistant Gram-negative bacteria in hospitalized patients. Clin Microbiol Infect 20(Suppl 1):1–55. doi: 10.1111/1469-0691.12427. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

