Spatial and temporal regularization to estimate COVID-19 reproduction number R(t): Promoting piecewise smoothness via convex optimization
- PMID: 32817697
- PMCID: PMC7444593
- DOI: 10.1371/journal.pone.0237901
Spatial and temporal regularization to estimate COVID-19 reproduction number R(t): Promoting piecewise smoothness via convex optimization
Abstract
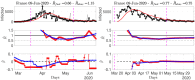
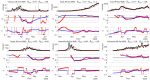
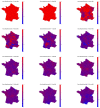
Among the different indicators that quantify the spread of an epidemic such as the on-going COVID-19, stands first the reproduction number which measures how many people can be contaminated by an infected person. In order to permit the monitoring of the evolution of this number, a new estimation procedure is proposed here, assuming a well-accepted model for current incidence data, based on past observations. The novelty of the proposed approach is twofold: 1) the estimation of the reproduction number is achieved by convex optimization within a proximal-based inverse problem formulation, with constraints aimed at promoting piecewise smoothness; 2) the approach is developed in a multivariate setting, allowing for the simultaneous handling of multiple time series attached to different geographical regions, together with a spatial (graph-based) regularization of their evolutions in time. The effectiveness of the approach is first supported by simulations, and two main applications to real COVID-19 data are then discussed. The first one refers to the comparative evolution of the reproduction number for a number of countries, while the second one focuses on French departments and their joint analysis, leading to dynamic maps revealing the temporal co-evolution of their reproduction numbers.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








References
-
- Petkova E, Antman EM, Troxel AB. Pooling Data From Individual Clinical Trials in the COVID-19 Era. JAMA. - PubMed
-
- Di Domenico L, Pullano G, Sabbatini CE, Boëlle PY, Colizza V. Expected impact of lockdown in Ile-de-France and possible exit strategies; 2020. medRxiv:2020.04.13.20063933. Available from: https://www.medrxiv.org/content/early/2020/04/17/2020.04.13.20063933. - PMC - PubMed
-
- Guzzetta et al G. The impact of a nation-wide lockdown on COVID-19 transmissibility in Italy; 2020. arXiv:2004.12338 [q-bio.PE].
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

