This is a preprint.
An ultra-potent synthetic nanobody neutralizes SARS-CoV-2 by locking Spike into an inactive conformation
- PMID: 32817938
- PMCID: PMC7430568
- DOI: 10.1101/2020.08.08.238469
An ultra-potent synthetic nanobody neutralizes SARS-CoV-2 by locking Spike into an inactive conformation
Update in
-
An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.Science. 2020 Dec 18;370(6523):1473-1479. doi: 10.1126/science.abe3255. Epub 2020 Nov 5. Science. 2020. PMID: 33154106 Free PMC article.
Abstract
Without an effective prophylactic solution, infections from SARS-CoV-2 continue to rise worldwide with devastating health and economic costs. SARS-CoV-2 gains entry into host cells via an interaction between its Spike protein and the host cell receptor angiotensin converting enzyme 2 (ACE2). Disruption of this interaction confers potent neutralization of viral entry, providing an avenue for vaccine design and for therapeutic antibodies. Here, we develop single-domain antibodies (nanobodies) that potently disrupt the interaction between the SARS-CoV-2 Spike and ACE2. By screening a yeast surface-displayed library of synthetic nanobody sequences, we identified a panel of nanobodies that bind to multiple epitopes on Spike and block ACE2 interaction via two distinct mechanisms. Cryogenic electron microscopy (cryo-EM) revealed that one exceptionally stable nanobody, Nb6, binds Spike in a fully inactive conformation with its receptor binding domains (RBDs) locked into their inaccessible down-state, incapable of binding ACE2. Affinity maturation and structure-guided design of multivalency yielded a trivalent nanobody, mNb6-tri, with femtomolar affinity for SARS-CoV-2 Spike and picomolar neutralization of SARS-CoV-2 infection. mNb6-tri retains stability and function after aerosolization, lyophilization, and heat treatment. These properties may enable aerosol-mediated delivery of this potent neutralizer directly to the airway epithelia, promising to yield a widely deployable, patient-friendly prophylactic and/or early infection therapeutic agent to stem the worst pandemic in a century.
Conflict of interest statement
Competing Interests M.Schoof, B.Faust, R.A.Saunders, N.Hoppe, P.Walter, and A.Manglik are inventors on a provisional patent describing anti-Spike nanobodies described in this manuscript.
Figures

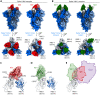

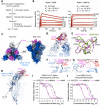
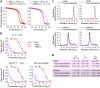
References
-
- Ksiazek T. G. et al., A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348, 1953–1966 (2003). - PubMed
-
- Zaki A. M., van Boheemen S., Bestebroer T. M., Osterhaus A. D., Fouchier R. A., Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367, 1814–1820 (2012). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
