This is a preprint.
Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2
- PMID: 32817941
- PMCID: PMC7430571
- DOI: 10.1101/2020.08.11.247395
Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2
Update in
-
Elicitation of Potent Neutralizing Antibody Responses by Designed Protein Nanoparticle Vaccines for SARS-CoV-2.Cell. 2020 Nov 25;183(5):1367-1382.e17. doi: 10.1016/j.cell.2020.10.043. Epub 2020 Oct 31. Cell. 2020. PMID: 33160446 Free PMC article.
Abstract
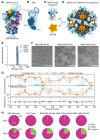
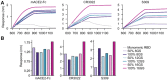
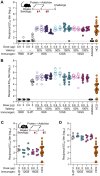
A safe, effective, and scalable vaccine is urgently needed to halt the ongoing SARS-CoV-2 pandemic. Here, we describe the structure-based design of self-assembling protein nanoparticle immunogens that elicit potent and protective antibody responses against SARS-CoV-2 in mice. The nanoparticle vaccines display 60 copies of the SARS-CoV-2 spike (S) glycoprotein receptor-binding domain (RBD) in a highly immunogenic array and induce neutralizing antibody titers roughly ten-fold higher than the prefusion-stabilized S ectodomain trimer despite a more than five-fold lower dose. Antibodies elicited by the nanoparticle immunogens target multiple distinct epitopes on the RBD, suggesting that they may not be easily susceptible to escape mutations, and exhibit a significantly lower binding:neutralizing ratio than convalescent human sera, which may minimize the risk of vaccine-associated enhanced respiratory disease. The high yield and stability of the protein components and assembled nanoparticles, especially compared to the SARS-CoV-2 prefusion-stabilized S trimer, suggest that manufacture of the nanoparticle vaccines will be highly scalable. These results highlight the utility of robust antigen display platforms for inducing potent neutralizing antibody responses and have launched cGMP manufacturing efforts to advance the lead RBD nanoparticle vaccine into the clinic.
Conflict of interest statement
DECLARATION OF INTERESTS A.C.W, D.V., and N.P.K. are named as inventors on patent applications filed by the University of Washington based on the studies presented in this paper. N.P.K. is a co-founder, shareholder, and chair of the scientific advisory board of Icosavax, Inc. H.Y.C. is a consultant for Merck and Pfizer, and has received research funding from Sanofi-Pasteur, Roche-Genentech, Cepheid, and Ellume outside of the submitted work. P.K., A.P., and S.C. are employees and shareholders of Kymab Ltd. The Veesler laboratory has received a sponsored research agreement from Vir Biotechnology Inc. The other authors declare no competing interests.
Figures



References
-
- Anywaine Z., Whitworth H., Kaleebu P., Praygod G., Shukarev G., Manno D., Kapiga S., Grosskurth H., Kalluvya S., Bockstal V., et al. (2019). Safety and Immunogenicity of a 2-Dose Heterologous Vaccination Regimen With Ad26.ZEBOV and MVA-BN-Filo Ebola Vaccines: 12-Month Data From a Phase 1 Randomized Clinical Trial in Uganda and Tanzania. J Infect Dis 220, 46–56. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
